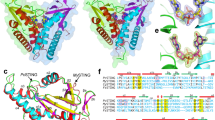
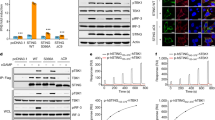
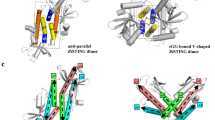
Abstract
Stimulator of interferon genes (STING) is a receptor in human cells that senses foreign cyclic dinucleotides that are released during bacterial infection and in endogenous cyclic GMP–AMP signalling during viral infection and anti-tumour immunity1,2,3,4,5. STING shares no structural homology with other known signalling proteins6,7,8,9, which has limited attempts at functional analysis and prevented explanation of the origin of cyclic dinucleotide signalling in mammalian innate immunity. Here we reveal functional STING homologues encoded within prokaryotic defence islands, as well as a conserved mechanism of signal activation. Crystal structures of bacterial STING define a minimal homodimeric scaffold that selectively responds to cyclic di-GMP synthesized by a neighbouring cGAS/DncV-like nucleotidyltransferase (CD-NTase) enzyme. Bacterial STING domains couple the recognition of cyclic dinucleotides with the formation of protein filaments to drive oligomerization of TIR effector domains and rapid NAD+ cleavage. We reconstruct the evolutionary events that followed the acquisition of STING into metazoan innate immunity, and determine the structure of a full-length TIR–STING fusion from the Pacific oyster Crassostrea gigas. Comparative structural analysis demonstrates how metazoan-specific additions to the core STING scaffold enabled a switch from direct effector function to regulation of antiviral transcription. Together, our results explain the mechanism of STING-dependent signalling and reveal the conservation of a functional cGAS–STING pathway in prokaryotic defence against bacteriophages.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Data that support the findings of this study are available within the Article, its Extended Data and Supplementary Information. IMG database accessions are listed in Extended Data Fig. 1, and PDB accessions are listed in each figure legend. Coordinates and structure factors of FsSTING–3′,3′-cGAMP, CgSTING, oyster TIR–STING, oyster TIR–STING–2′,3′-cGAMP, FsCdnE and CgCdnE have been deposited in PDB under accession codes 6WT4, 6WT5, 6WT6, 6WT7, 6WT8 and 6WT9, respectively. Source data are provided with this paper.
References
Ishikawa, H. & Barber, G. N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678 (2008).
Zhong, B. et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29, 538–550 (2008).
Burdette, D. L. et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518 (2011).
Sun, L., Wu, J., Du, F., Chen, X. & Chen, Z. J. Cyclic GMP–AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013).
Ablasser, A. & Chen, Z. J. cGAS in action: expanding roles in immunity and inflammation. Science 363, eaat8657 (2019).
Ouyang, S. et al. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity 36, 1073–1086 (2012).
Zhang, X. et al. Cyclic GMP–AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol. Cell 51, 226–235 (2013).
Kranzusch, P. J. et al. Ancient origin of cGAS–STING reveals mechanism of universal 2′,3′ cGAMP signaling. Mol. Cell 59, 891–903 (2015).
Shang, G., Zhang, C., Chen, Z. J., Bai, X. C. & Zhang, X. Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP–AMP. Nature 567, 389–393 (2019).
Cohen, D. et al. Cyclic GMP–AMP signalling protects bacteria against viral infection. Nature 574, 691–695 (2019).
Gui, X. et al. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature 567, 262–266 (2019).
Zhang, C. et al. Structural basis of STING binding with and phosphorylation by TBK1. Nature 567, 394–398 (2019).
Zhao, B. et al. A conserved PLPLRT/SD motif of STING mediates the recruitment and activation of TBK1. Nature 569, 718–722 (2019).
de Oliveira Mann, C. C. et al. Modular architecture of the STING C-terminal tail allows interferon and NF-κB signaling adaptation. Cell Rep. 27, 1165–1175 (2019).
Whiteley, A. T. et al. Bacterial cGAS-like enzymes synthesize diverse nucleotide signals. Nature 567, 194–199 (2019).
Ye, Q. et al. HORMA domain proteins and a Trip13-like ATPase regulate bacterial cGAS-like enzymes to mediate bacteriophage immunity. Mol. Cell 77, 709–722 (2020).
Lowey, B. et al. CBASS immunity uses CARF-related effectors to sense 3′-5′- and 2'-5′-linked cyclic oligonucleotide signals and protect bacteria from phage infection. Cell 182, 38–49 (2020).
Jenal, U., Reinders, A. & Lori, C. Cyclic di-GMP: second messenger extraordinaire. Nat. Rev. Microbiol. 15, 271–284 (2017).
Gao, P. et al. Structure-function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell 154, 748–762 (2013).
Essuman, K. et al. TIR domain proteins are an ancient family of NAD+-consuming enzymes. Curr. Biol. 28, 421–430 (2018).
Horsefield, S. et al. NAD+ cleavage activity by animal and plant TIR domains in cell death pathways. Science 365, 793–799 (2019).
Wan, L. et al. TIR domains of plant immune receptors are NAD+-cleaving enzymes that promote cell death. Science 365, 799–803 (2019).
Ergun, S. L., Fernandez, D., Weiss, T. M. & Li, L. STING polymer structure reveals mechanisms for activation, hyperactivation, and inhibition. Cell 178, 290–301 (2019).
Margolis, S. R., Wilson, S. C. & Vance, R. E. Evolutionary origins of cGAS–STING signaling. Trends Immunol. 38, 733–743 (2017).
Zhang, G. et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49–54 (2012).
Toshchakov, V. Y. & Neuwald, A. F. A survey of TIR domain sequence and structure divergence. Immunogenetics 72, 181–203 (2020).
Wexler, A. G. & Goodman, A. L. An insider’s perspective: Bacteroides as a window into the microbiome. Nat. Microbiol. 2, 17026 (2017).
Woodward, J. J., Iavarone, A. T. & Portnoy, D. A. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328, 1703–1705 (2010).
Dey, B. et al. A bacterial cyclic dinucleotide activates the cytosolic surveillance pathway and mediates innate resistance to tuberculosis. Nat. Med. 21, 401–406 (2015).
Sixt, B. S. et al. The Chlamydia trachomatis inclusion membrane protein CpoS counteracts STING-mediated cellular surveillance and suicide programs. Cell Host Microbe 21, 113–121 (2017).
Burroughs, A. M., Zhang, D., Schäffer, D. E., Iyer, L. M. & Aravind, L. Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling. Nucleic Acids Res. 43, 10633–10654 (2015).
Loring, H. S., Icso, J. D., Nemmara, V. V. & Thompson, P. R. Initial kinetic characterization of sterile alpha and Toll/interleukin receptor motif-containing protein 1. Biochemistry 59, 933–942 (2020).
Tak, U. et al. The tuberculosis necrotizing toxin is an NAD+ and NADP+ glycohydrolase with distinct enzymatic properties. J. Biol. Chem. 294, 3024–3036 (2019).
Ghosh, J., Anderson, P. J., Chandrasekaran, S. & Caparon, M. G. Characterization of Streptococcus pyogenes β-NAD+ glycohydrolase: re-evaluation of enzymatic properties associated with pathogenesis. J. Biol. Chem. 285, 5683–5694 (2010).
Millman, A., Melamed, S., Amitai, G. & Sorek, R. Diversity and classification of cyclic-oligonucleotide-based anti-phage signalling systems. Nat. Microbiol. https://doi.org/10.1038/s41564-020-0777-y (2020).
Chen, I. A. et al. IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 47, D666–D677 (2019).
Zhou, W. et al. Structure of the human cGAS–DNA complex reveals enhanced control of immune surveillance. Cell 174, 300–311 (2018).
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Huang, Y. H., Liu, X. Y., Du, X. X., Jiang, Z. F. & Su, X. D. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nat. Struct. Mol. Biol. 19, 728–730 (2012).
Shang, G. et al. Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nat. Struct. Mol. Biol. 19, 725–727 (2012).
Shu, C., Yi, G., Watts, T., Kao, C. C. & Li, P. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat. Struct. Mol. Biol. 19, 722–724 (2012).
Yin, Q. et al. Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol. Cell 46, 735–745 (2012).
Cavlar, T., Deimling, T., Ablasser, A., Hopfner, K. P. & Hornung, V. Species-specific detection of the antiviral small-molecule compound CMA by STING. EMBO J. 32, 1440–1450 (2013).
Zhang, H. et al. Rat and human STINGs profile similarly towards anticancer/antiviral compounds. Sci. Rep. 5, 18035 (2015).
Cong, X. et al. Crystal structures of porcine STINGCBD–CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins. J. Biol. Chem. 294, 11420–11432 (2019).
Krissinel, E. & Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D 60, 2256–2268 (2004).
Pei, J. & Grishin, N. V. PROMALS3D: multiple protein sequence alignment enhanced with evolutionary and three-dimensional structural information. Methods Mol. Biol. 1079, 263–271 (2014).
Katoh, K., Rozewicki, J. & Yamada, K. D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166 (2019).
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M. & Barton, G. J. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019).
Kulasakara, H. et al. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl Acad. Sci. USA 103, 2839–2844 (2006).
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Kranzusch, P. J. et al. Structure-guided reprogramming of human cGAS dinucleotide linkage specificity. Cell 158, 1011–1021 (2014).
Acknowledgements
We thank J. Morehouse, A. Lee, K. Chat, R. Vance and members of the Kranzusch laboratory for helpful comments and discussion; K. Arnett and the Harvard University Center for Macromolecular Interactions; the Molecular Electron Microscopy Suite at Harvard Medical School; and the Harvard Center for Mass Spectrometry. The work was funded by the Richard and Susan Smith Family Foundation (P.J.K. and S.S.), DFCI-Novartis Drug Discovery Program (P.J.K.), the Parker Institute for Cancer Immunotherapy (P.J.K.), a Cancer Research Institute CLIP Grant (P.J.K.), a V Foundation V Scholar Award (P.J.K.), the Pew Biomedical Scholars program (P.J.K.), Vallee Foundation (S.S.), the Ariane de Rothschild Women Doctoral Program (A.M.), the Israeli Council for Higher Education via the Weizmann Data Science Research Center (A.M.), the European Research Council (grant ERC-CoG 681203 to R.S.), the Ernest and Bonnie Beutler Research Program of Excellence in Genomic Medicine (R.S.), the Minerva Foundation (R.S.) and the Knell Family Center for Microbiology (R.S.). B.R.M. is supported as a Ruth L. Kirschstein NRSA Postdoctoral Fellow NIH F32GM133063, A.A.G. is supported by a United States National Science Foundation Graduate Research Fellowship, B.L. is supported as a Herchel Smith Graduate Research Fellow, G.O. is supported by a Weizmann Sustainability and Energy Research Initiative (SAERI) doctoral fellowship. X-ray data were collected at the Northeastern Collaborative Access Team beamlines 24-ID-C and 24-ID-E (P30 GM124165), and used a Pilatus detector (S10RR029205), an Eiger detector (S10OD021527) and the Argonne National Laboratory Advanced Photon Source (DE-AC02-06CH11357).
Author information
Authors and Affiliations
Contributions
Experiments were designed and conceived by B.R.M., R.S. and P.J.K. Structural and biochemical experiments were performed by B.R.M. with assistance from A.A.G. and P.J.K. NAD+ cleavage assays were performed by A.A.G. and B.R.M. Gene identification and phylogenetic analysis were performed by A.M. and R.S. Electron microscopy experiments and analysis were conducted by A.F.A.K. and S.S. STING oligomerization analysis was performed by B.L. and B.R.M. STING toxicity analysis was performed by G.O. and R.S. The manuscript was written by B.R.M. and P.J.K. All authors contributed to editing the manuscript, and support the conclusions.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Urs Jenal and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Structural analysis of bacterial STING–cyclic dinucleotide complex formation.
a, Phylogenetic tree of all CBASS-associated bacterial STING homologues based on structure-guided sequence alignment and previous bioinformatics analysis10,35. STING homologues investigated in this study are highlighted in orange, and a star denotes determined STING crystal structures. All TM–STING fusions cluster together. b, Crystal structure of a STING receptor from the bacterium C. granulosa (CgSTING) in the apo state reveals an open configuration with a solvent exposed cyclic-dinucleotide-binding pocket at the dimeric interface (monomers in gold and grey for clarity). The CgSTING structure confirms that both divergent TM–STING and TIR–STING fusions are members of the same structurally conserved family of STING receptors. c, Comparison of the CgSTING, FsSTING–3′,3′-cGAMP and human STING–2′,3′-cGAMP structures demonstrates conservation of an open-to-closed β-strand lid movement upon ligand binding. d, Overlay of the β-strand lid of CgSTING (grey) and FsSTING (orange) shows both inward translation and slight rotation resulting in a displacement of about 5 Å. R153 of FsSTING stacks between the bases of 3′,3′-cGAMP and R151 is splayed away from ligand. e, Comparison of the human STING and FsSTING lid region shows conserved contacts from β-strand arginine residues. Unlike in bacterial STING, human STING R232 makes an additional contact with the cyclic dinucleotide phosphodiester backbone that is critical for recognition of the 2′–5′ linkage in 2′,3′-cGAMP. A detailed comparison is in Extended Data Fig. 10b. f, Modelling of 2′,3′-cGAMP into the FsSTING–3′,3′-cGAMP structure demonstrates an additional feature of bacterial STING preventing recognition of 2′–5′-linked cyclic dinucleotides. Although the overall cyclic dinucleotide conformation is shared between human and bacterial STING, the α-helix ending at P264 in human STING is a half-turn longer in FsSTING (also ending in a proline) which places a conserved T173 residue in a position that occludes where the free 3′-OH of 2′,3′-cGAMP would be positioned. g, Structure-guided alignment of FsSTING and human STING cyclic-dinucleotide-binding domains. FsSTING and human STING exhibit no detectable sequence homology but share a conserved structural fold. Key residues involved in cyclic dinucleotide binding that are shared between bacterial and human STING are boxed in orange and human STING specific cyclic dinucleotide contacts are boxed in red. In FsSTING, D169 directly reads out the guanine base of c-di-GMP.
Extended Data Fig. 2 Structural analysis of STING-associated CD-NTase enzymes.
a, Sequence and secondary structure alignment of STING-associated CD-NTases reveals the extent of homology between CdnE homologues from unrelated bacterial strains. Highlighted positions include active-site residues (pink box), and an aspartic acid substitution at a position known to be involved in nucleotide substrate selection (orange box) that is unique to CD-NTases in STING-containing CBASS operons15. A divergent E. coli CdnE that synthesizes cyclic UMP–AMP is included for comparison. b, Crystal structures of FsCdnE and CgCdnE from STING-containing CBASS operons allow direct comparison with previously determined bacterial and human CD-NTase structures. The FsCdnE and CgCdnE structures are most closely related to the clade-E CD-NTase structure from Rhodothermus marinus CdnE (RmCdnE). RmCdnE: PDB 6E0L15; V. cholerae DncV: PDB 4TY057; and human cGAS: PDB 6CTA (DNA omitted for clarity)37.
Extended Data Fig. 3 Biochemical analysis of c-di-GMP synthesis by bacterial STING-associated CD-NTases.
a, In addition to FsCdnE (Fig. 2b), CdnE homologues from three divergent STING-containing CBASS operons were purified and tested for cyclic-dinucleotide-synthesis specificity using α32P-radiolabelled NTPs and thin-layer chromatography. Deconvolution experiments show a single major product requiring only GTP that migrates identically to c-di-GMP synthesized by the GGDEF enzyme WspR. All reactions were treated with alkaline phosphatase to remove exposed phosphates. Only two bacterial genomes encoding a STING-containing CBASS operon retain proteins with a predicted canonical GGDEF c-di-GMP signalling domain. The exceptions are chlorobi bacterium EBPR_Bin_190, which contains a single GGDEF domain that is fused to a SLATT domain and may be part of a CBASS-like system10, and a Lachnospiraceae bacterium RUG226 genome that encodes many GGDEF genes. The Lachnospiraceae bacterium RUG226 CdnE retains exclusive production of c-di-GMP suggesting the CdnE–STING system is sequestered in this bacterium or that an unknown mechanism may exist to prevent toxic STING activation. ReCdnE, Roseivirga ehrenbergii; CgCdnE, Capnocytophaga granulosa; LbCdnE, Lachnospiraceae bacterium; N, all four rNTPs; Pi, inorganic phosphate; ori., origin. Data are representative of two independent experiments. b, Nuclease treatment confirms that the FsCdnE enzymatic product contains only canonical 3′–5′ phosphodiester bonds. The [α32P]GTP cyclic dinucleotide product is susceptible to cleavage by nuclease P1 resulting in release of GMP as a new species, which migrates further up the TLC plate. Further digestion with calf-intestinal phosphatase (CIP) removes all exposed phosphates, resulting in complete loss of a labelled product spot. DncV (Dinucleotide cyclase in Vibrio)-derived 3′,3′-cGAMP is similarly susceptible to complete digestion by P1 and CIP treatment, whereas 2′,3′-cGAMP synthesized by mouse cGAS is only partially digested owing to the presence of the non-canonical 2′–5′ bond. Data are representative of two independent experiments. c, High-resolution mass spectrometry analysis confirms the identity of the major FsCdnE enzymatic product as canonical c-di-GMP. Chemically synthesized c-di-GMP was used for direct spectral comparison. d, Sequence alignment and enlarged inset of the active-site of the RmCdnE structure in complex with nonhydrolyzable UTP and ATP analogues (PDB 6E0L), demonstrating a contact in the CD-NTase lid domain known to control nucleobase sequence specificity15. RmCdnE synthesizes cyclic UMP–AMP and uses N166 to specifically contact the uridine Watson–Crick edge. By contrast, FsCdnE and CgCdnE contain an aspartic acid substitution at this position and synthesize c-di-GMP, and V. cholerae DncV and human cGAS contain a serine substitution at this position and synthesize 3′,3′-cGAMP and 2′,3′-cGAMP. An aspartic acid at the FsCdnE D233 position is conserved among 93% of STING-associated CD-NTase enzymes (96 of 103), consistent with strict specificity of c-di-GMP as the nucleotide second messenger that controls bacterial STING activation. RmCdnE: PDB 6E0L15; V. cholerae DncV: PDB 4TY057; and human cGAS: PDB 6CTA (DNA omitted for clarity)37. e, f, Mutational analysis of the importance of D233 in FsCdnE c-di-GMP synthesis activity. D233 substitutions do not disrupt the overall ability of FsCdnE to selectively synthesis c-di-GMP, but a D233A substitution causes a mild reduction in nucleobase selectivity and efficiency of c-di-GMP synthesis. These results are consistent with a role for D233 in nucleobase selection but demonstrate full selectively is achieved by additional contacts in the active site pocket. Data are representative of three independent experiments.
Extended Data Fig. 4 Biochemical analysis of bacterial STING cyclic dinucleotide recognition specificity.
a, b, Electrophoretic mobility shift assay (EMSA) of purified bacterial STING proteins with radiolabelled cyclic dinucleotide ligands. Bacterial STING receptors specifically recognize c-di-GMP and have a weak ability to bind 3′,3′-cGAMP. No interaction was observed with c-di-AMP or 2′,3′-cGAMP. Lachnospiraceae bacterium STING: LbSTING; Aggregatibacter actinomycetemcomitans STING, AaSTING. Data are representative of two independent experiments. c, EMSA analysis of a diverse panel of bacterial STING homologues demonstrates conservation of c-di-GMP binding in both TM–STING and TIR–STING CBASS immunity. NdSTING, Niabella drilacis; FdSTING, Flavobacterium daejeonense. Higher-order complex formation visible as well-shifted complexes is consistent with STING oligomerization results (Fig. 3f, Extended Data Fig. 7). Data are representative of three independent experiments. d, EMSA analysis of diverse bacterial STING homologues broadly demonstrates no interaction with the 3′,3′-c-UMP–AMP second messenger synthesized by the divergent CD-NTase E. coli CdnE15 and further confirms the specificity of c-di-GMP signalling in bacterial STING-containing CBASS operons. Data are representative of two independent experiments. e–h, EMSA analysis and quantification of the affinity of bacterial STING homologues for c-di-GMP and 3′,3′-cGAMP. Signal intensity analysis is plotted as fraction bound (shifted/total signal) as a function of increasing protein concentration and fit to a single binding isotherm. CgSTING and ReSTING have a >10-fold preference for c-di-GMP whereas SfSTING has a similar apparent affinity for c-di-GMP and 3′,3′-cGAMP. Data are representative of two independent experiments.
Extended Data Fig. 5 Bacterial STING activation of TIR NADase activity.
a, HPLC analysis of chemical standards separated with an ammonium acetate:methanol gradient elution used to analyse bacterial TIR–STING activity (Methods). The NAD+ and ADPr peaks have overlapping bases under these conditions. cADPr, cyclic adenosine diphosphate-ribose; ADPr, adenosine diphosphate-ribose; NAD+, β-nicotinamide adenine dinucleotide; NAM, nicotinamide. b, HPLC analysis of SfSTING NAD+ cleavage activity with gradient elution. SfSTING at 500 nM protein with 2 μM c-di-GMP converts 500 μM NAD+ into ADPr and NAM in 30 min at ambient temperature. SfSTING does not generate any cyclized product and is therefore a standard glycosyl hydrolase. Inset, schematic of NAD+ cleavage reaction. c, HPLC analysis of chemical standards separated with an alternative isocratic elution strategy (Methods) that results in clearer separation of NAD+ and ADPr peaks. d, HPLC analysis of SfSTING NAD+ cleavage activity with isocratic elution. SfSTING NAD+ cleavage activity requires specific activation with c-di-GMP (30-min reactions). e, HPLC analysis of SfSTING NAD+ cleavage activity and cyclic dinucleotide agonist specificity. Each reaction was tested with 500 nM SfSTING, 500 μM cyclic dinucleotide and 500 μM NAD+ and sampled at 45, 90 or 180 min (gradient colouring in bars). SfSTING preferentially responds to c-di-GMP, but 3′,3′-cGAMP and c-di-AMP can function as weak agonists. Data are representative of three independent experiments. f, HPLC analysis of SfSTING NAD+ cleavage activity in the presence of 3′,3′-c-UMP–AMP. 3′,3′-c-UMP–AMP is a >1,000×-weaker agonist than c-di-GMP. Data are representative of three independent experiments. g, HPLC analysis of SfSTING NAD+ cleavage activity in the presence of a synthetic c-di-GMP analogue with a noncanonical 2′–5′ linkage (2′,3′-c-di-GMP). 2′,3′-c-di-GMP is not capable of stimulating robust SfSTING activation even at very high concentrations (250 μM versus 250 nM canonical c-di-GMP), confirming the specificity of bacterial STING for 3′–5′-linked cyclic dinucleotides. Data are representative of three independent experiments. h, Plate reader analysis of SfSTING NAD+ cleavage activity using the fluorescent substrate ε-NAD. ε-NAD increases in fluorescence intensity after cleavage. SfSTING exhibits rapid catalysis with complete turnover at 500 nM protein with 500 nM c-di-GMP after 10 min at 25 °C. No background activity is observed in the absence of ligand. Data are representative of three independent experiments. i, Plate reader analysis of SfSTING NAD+ cleavage activity in the presence of 500 nM protein with increasing c-di-GMP concentration reveals that low nM c-di-GMP levels are sufficient to induce ε-NAD cleavage. Greater c-di-GMP concentrations are required for maximal activity, consistent with binding data and the higher amount of c-di-GMP required for complete stabilization of the SfSTING–c-di-GMP complex (Extended Data Fig. 4e–h). Saturation occurs above 100 nM c-di-GMP with 40-min reactions. Data are ± s.d. of n = 3 technical replicates and are representative of 3 independent experiments. j, k, TIR-domain NAD+ cleavage activity requires protein oligomerization. For other systems21,22, TIR activation has been observed at very high in vitro protein concentrations or in the presence of affinity resins as an artificial oligomerization-inducing matrix21,22. We used a GST–TIR construct to express the SfSTING TIR domain in absence of the STING cyclic-dinucleotide-binding domain and observed that even at >200× the concentrations for which the full-length protein shows c-di-GMP induced activity, or in the presence of multivalent affinity resin, no NAD+ cleavage activity occurs. These results demonstrate NAD+ cleavage activity specifically requires STING cyclic dinucleotide recognition for activation. Data are representative of two independent experiments.
Extended Data Fig. 6 Mutagenesis analysis of bacterial STING cyclic dinucleotide recognition and TIR activation.
a, Table of bacterial STING cyclic dinucleotide contacts tested with mutagenesis analysis. Residues were selected according to contacts observed in the structure of the FsSTING–3′,3′-cGAMP complex (FsSTING residues listed in parentheses) and tested in SfSTING to allow analysis of the effect on both c-di-GMP binding and NADase activity. b, Plate reader analysis of mutant SfSTING NAD+ cleavage activity in the presence of 500 nM protein and increasing c-di-GMP concentration. Mutant SfSTING variants with cyclic-dinucleotide-binding pocket mutations require 10–1,000× greater c-di-GMP concentration for NADase activation. Data are ± s.d. of n = 3 technical replicates and are representative of 3 independent experiments. c–e, EMSA analysis demonstrating that SfSTING R234A and D259A mutations reduce stable c-di-GMP complex formation compared to SfSTING wild type (Fig. 2d). SfSTING(D259A) protein titration and quantification confirms significantly reduced affinity for c-di-GMP. Data are representative of three independent experiments. f, g, HPLC and plate reader analysis of mutant SfSTING NAD+ cleavage activity in the presence of 500 nM protein and 10 μM c-di-GMP (HPLC) or 500 nM protein and ± 20 μM c-di-GMP (plate reader). Mutation of residues responsible for ligand recognition in the cyclic-dinucleotide-binding domain (R234 and D259) and catalysis in the TIR enzymatic domain (E84) disrupts SfSTING NADase activity and explains loss of E. coli toxicity observed in Fig. 3d. One hundred μM of c-di-GMP was used for SfSTING(D259A) plate reader NAD+ cleavage analysis to confirm complete loss of c-di-GMP-induced activation. HPLC data are representative of three independent experiments. Plate reader data are ± s.d. of n = 3 technical replicates and are representative of 3 independent experiments. h, i, Analysis of SfSTING toxicity in E. coli cells expressing normal c-di-GMP signalling enzymes with and without nicotinamide (NAM) supplementation. NAM supplementation is sufficient to partially alleviate SfSTING-wild-type-induced NADase toxicity. Each line represents the average of two technical replicates for each of four separately outgrown colonies. Data are representative of two independent experiments.
Extended Data Fig. 7 Conservation of oligomerization as a mechanism of STING activation.
a–i, STING SEC-MALS analysis. a, b, Full-length SfSTING changes oligomeric state in the presence of c-di-GMP or the weak agonist 3′,3′-cGAMP, and does not change oligomeric state in the presence of 2′,3′-cGAMP. c, With the TIR domain removed (ΔTIR), SfSTING no longer forms higher-order complexes but notably remains dimeric in the apoprotein form. Bacterial TIR–STING proteins therefore appear to require the TIR domain to maintain stable higher-order oligomerization, suggesting that intermolecular contacts are made with both TIR and STING domains. d, A TIR-only construct of SfSTING with the cyclic-dinucleotide-binding domain removed (ΔCDN) elutes as a single species that is consistent with the molecular weight for a homodimer. h, Human STING (ΔTM) is a dimer in solution with or without 2′,3′-cGAMP, confirming that transmembrane contacts are required for oligomerization and filament formation12,23. Nearly all tested bacterial and metazoan STING constructs migrate as dimers in solution, consistent with the cyclic-dinucleotide-binding domain forming a constitutive homodimeric complex for ligand recognition. Two exceptions include ReSTING(ΔTM) and FsSTING(ΔTM), which form a mixture of monomeric and dimeric states in the absence of ligand and dimers or tetramers in the presence of 3′,3′-cGAMP. These results indicate that alternative oligomerization events may be required for activation of bacterial TM–STING effector function. j, Negative-stain electron microscopy 2D class averages for SfSTING (E84A mutant) alone or in the presence of cyclic dinucleotide ligands. Stable STING filament formation requires c-di-GMP. Two-dimensional class averages were derived from particles selected from 75 micrographs for each condition. k, Representative micrograph images reveal extensive filament formation of varying length and orientation in the presence of c-di-GMP. Apo, c-di-AMP and 2′,3′-cGAMP micrographs lack filaments. Images are each representative of n = 75 micrographs for each condition. l, Particle counting analysis of micrograph images shows that c-di-GMP induces more filament formation than 3′,3′-cGAMP, and stable filament formation does not occur in the presence of c-di-AMP or 2′,3′-cGAMP. Data are mean ± s.d. for quantification of n = 4 groups of 10 micrograph images each.
Extended Data Fig. 8 Filament formation is required for bacterial TIR–STING activation.
a, Model of bacterial STING oligomerization and identification of surfaces involved in c-di-GMP-mediated filament formation. Electron microscopy analysis of SfSTING in the presence of c-di-GMP reveals filament formation probably occurs through parallel stacking of the homodimeric cyclic-dinucleotide-binding domain (Extended Data Fig. 7). To construct a potential model of this interaction, we used the X-ray crystal structure of the human STING–2′,3′-cGAMP complex (PDB 4KSY)7 and the cryo-electron microscopy structure of the chicken STING tetramer (PDB 6NT8)9 (top) as guides to position the FsSTING–3′,3′-cGAMP complex structure into a tetrameric conformation. The resulting model predicts that oligomerization-mediating surfaces in SfSTING include T200–N204, L275–F284 and G306–A310. b, EMSA analysis of SfSTING variants indicates that mutations to the predicted oligomerization surfaces do not prevent c-di-GMP recognition. SfSTING mutants tested include R307E/A309R, L201R/D203R and the loop L275–Q282 replaced with a short GlySer linker (GSGGS). Data are representative of two independent experiments. c, Electron microscopy analysis of SfSTING variants in the presence of c-di-GMP. Mutations to the SfSTING surfaces identified in the structural model prevent all observable cyclic-dinucleotide-induced filament formation, supporting their predicted role in mediating oligomerization and bacterial STING filament formation. Images are each representative of n = 5 micrographs for each condition. d, HPLC analysis of mutant SfSTING NAD+ cleavage activity in the presence of 500 nM protein and 250 μM c-di-GMP for 3 h. In the absence of cyclic-dinucleotide-mediated oligomerization, all SfSTING NADase activity is lost, confirming the requirement of filament formation in TIR domain activation. Data are representative of three independent experiments.
Extended Data Fig. 9 Structural analysis of metazoan TIR–STING homologues.
a, Structure-guided alignment of the TIR domain in oyster TIR–STING with reference bacterial and metazoan TIR-domain-containing proteins. SARM1 is an example of a human TIR domain that catalyses NAD+ cleavage, and MyD88 is an example of a human TIR domain that signals through protein–protein interaction. The catalytic glutamate responsible for supporting NAD+ cleavage is conserved at the same spatial position among bacterial and oyster TIRs but is mutated in MyD88 (green box). However, it is not currently possible to predict from structure or sequence alone whether TIR domains have enzymatic activity. b, Distinct from other TIR domain structures, the TIR domain in oyster TIR–STING contains a proline-rich loop region at the interface, suggesting a specific role in dimer stabilization. c, Superposition of a homology model of the SfSTING TIR domain with the TIR domain of oyster TIR–STING shows the predicted catalytic glutamates for both proteins occupy distinct locations in the TIR fold. d, Superposition of a homology model of the SfSTING TIR domain compared to the crystal structure of human SARM1 bound to ribose implies that different NAD+ binding pockets may exist between bacterial and eukaryotic TIRs, as previously suggested21. e, Superposition of a homology model of the SfSTING TIR domain with the bacterial TIR domain from Paracoccus denitrificans shows a high degree of similarity. No crystal structures are available for bacterial TIR domains in an active state, preventing identification of a specific mechanism of catalytic activation. f, EMSA analysis of oyster TIR–STING and mouse STING demonstrates a wide preference for cyclic dinucleotide interactions and clear ability to recognize the mammalian cGAS product 2′,3′-cGAMP. Data are representative of three independent experiments. g, h, Oyster TIR–STING, which binds all tested cyclic dinucleotides, does not exhibit NAD+ cleavage activity even at 10× the protein and ligand concentrations used to achieve robust activity with bacterial TIR–STING. We tested four other oyster TIR–STING homologues and observed no cyclic-dinucleotide-stimulated NAD+ cleavage activity. These results support a potential switch in TIR-dependent protein–protein interactions to control downstream signalling similar to the TIR domain in human MyD88. Data are representative of two independent experiments.
Extended Data Fig. 10 Structure-guided analysis of STING phylogenetic conservation and cyclic dinucleotide recognition.
a, Structure-guided alignment and phylogenetic tree of STING proteins across bacterial and metazoan kingdoms. Bacterial STING homologues form a distinct cluster separate from all metazoan STING sequences, and are mostly represented by TIR–STING fusions. A single STING-domain containing protein was identified in the choanoflagellate Monosiga brevicollis (denoted as an open black circle, as this species is outside of the kingdom Metazoa); no STING-domain containing proteins were found in Archaea. TIR–STING fusions are rare in eukaryotes and cluster among invertebrate metazoans. No TIR–STING examples occur in vertebrates. Specific species of interest are highlighted to show the breadth of sequence diversity and stars mark proteins with available structures. b, Direct comparison of bacterial, oyster, anemone and human STING crystal structures reveals conservation of specific cyclic dinucleotide contacts and critical differences in phosphodiester linkage recognition. Stacking interactions formed with the cyclic dinucleotide nucleobase face, aromatic side chains at the top of the α-helix stem and arginine residues extending downward from the lid region are major conserved features shared between bacterial and metazoan STING proteins. Nucleotide-specific contacts are divergent between distinct STING structures, but notably the critical D169 guanosine N2-specific contact present in bacterial STING is conserved with a glutamic acid side chain in nearly all metazoan STING proteins. A critical feature absent in bacterial STING receptors is additional arginine-specific contacts to the phosphodiester backbone. The human STING R232 side chain contact, known to be critical for high-affinity interactions with 2′,3′-cGAMP, is conserved throughout metazoan STINGs, representing a unique adaptation not found within bacterial STING receptors. Bacterial STING–3′,3′-cGAMP (FsSTING), oyster STING–2′,3′-cGAMP (C. gigas), sea anemone STING–2′,3′-cGAMP (N. vectensis PDB 5CFQ)8, human STING–2′,3′-cGAMP (H. sapiens PDB 4KSY)7.
Supplementary information
Supplementary Table 1
This table contains a summary of X-ray data collection, phasing and refinement statistics.
Rights and permissions
About this article
Cite this article
Morehouse, B.R., Govande, A.A., Millman, A. et al. STING cyclic dinucleotide sensing originated in bacteria. Nature 586, 429–433 (2020). https://doi.org/10.1038/s41586-020-2719-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2719-5
This article is cited by
-
Nanomaterial-encapsulated STING agonists for immune modulation in cancer therapy
Biomarker Research (2024)
-
Reversible conjugation of a CBASS nucleotide cyclase regulates bacterial immune response to phage infection
Nature Microbiology (2024)
-
STING agonist diABZI enhances the cytotoxicity of T cell towards cancer cells
Cell Death & Disease (2024)
-
Auto-inhibition and activation of a short Argonaute-associated TIR-APAZ defense system
Nature Chemical Biology (2024)
-
Conservation and similarity of bacterial and eukaryotic innate immunity
Nature Reviews Microbiology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.