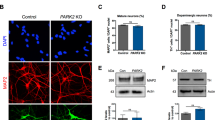
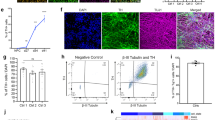
Abstract
Nuclear-architecture defects have been shown to correlate with the manifestation of a number of human diseases as well as ageing1,2,3,4. It is therefore plausible that diseases whose manifestations correlate with ageing might be connected to the appearance of nuclear aberrations over time. We decided to evaluate nuclear organization in the context of ageing-associated disorders by focusing on a leucine-rich repeat kinase 2 (LRRK2) dominant mutation (G2019S; glycine-to-serine substitution at amino acid 2019), which is associated with familial and sporadic Parkinson’s disease as well as impairment of adult neurogenesis in mice5. Here we report on the generation of induced pluripotent stem cells (iPSCs) derived from Parkinson’s disease patients and the implications of LRRK2(G2019S) mutation in human neural-stem-cell (NSC) populations. Mutant NSCs showed increased susceptibility to proteasomal stress as well as passage-dependent deficiencies in nuclear-envelope organization, clonal expansion and neuronal differentiation. Disease phenotypes were rescued by targeted correction of the LRRK2(G2019S) mutation with its wild-type counterpart in Parkinson’s disease iPSCs and were recapitulated after targeted knock-in of the LRRK2(G2019S) mutation in human embryonic stem cells. Analysis of human brain tissue showed nuclear-envelope impairment in clinically diagnosed Parkinson’s disease patients. Together, our results identify the nucleus as a previously unknown cellular organelle in Parkinson’s disease pathology and may help to open new avenues for Parkinson’s disease diagnoses as well as for the potential development of therapeutics targeting this fundamental cell structure.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Liu, G. H. et al. Recapitulation of premature ageing with iPSCs from Hutchinson–Gilford progeria syndrome. Nature 472, 221–225 (2011)
Dechat, T. et al. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 22, 832–853 (2008)
Kudlow, B. A., Kennedy, B. K. & Monnat, R. J., Jr Werner and Hutchinson–Gilford progeria syndromes: mechanistic basis of human progeroid diseases. Nature Rev. Mol. Cell Biol. 8, 394–404 (2007)
Worman, H. J., Ostlund, C. & Wang, Y. Diseases of the nuclear envelope. Cold Spring Harb. Perspect. Biol. 2, a000760 (2010)
Winner, B. et al. Adult neurogenesis and neurite outgrowth are impaired in LRRK2 G2019S mice. Neurobiol. Dis. 41, 706–716 (2011)
Chang, K. H. et al. Nuclear envelope dispersion triggered by deregulated Cdk5 precedes neuronal death. Mol. Biol. Cell 22, 1452–1462 (2011)
Tran, D., Chalhoub, A., Schooley, A., Zhang, W. & Ngsee, J. K. A mutation in VAPB that causes amyotrophic lateral sclerosis also causes a nuclear envelope defect. J. Cell Sci. 125, 2831–2836 (2012)
Padiath, Q. S. et al. Lamin B1 duplications cause autosomal dominant leukodystrophy. Nature Genet. 38, 1114–1123 (2006)
Woulfe, J. M. Abnormalities of the nucleus and nuclear inclusions in neurodegenerative disease: a work in progress. Neuropathol. Appl. Neurobiol. 33, 2–42 (2007)
Cookson, M. R. The role of leucine-rich repeat kinase 2 (LRRK2) in Parkinson's disease. Nature Rev. Neurosci. 11, 791–797 (2010)
Cookson, M. R. & Bandmann, O. Parkinson's disease: insights from pathways. Hum. Mol. Genet. 19, R21–R27 (2010)
Deng, X. et al. Characterization of a selective inhibitor of the Parkinson's disease kinase LRRK2. Nature Chem. Biol. 7, 203–205 (2011)
Lee, B. D. et al. Inhibitors of leucine-rich repeat kinase-2 protect against models of Parkinson's disease. Nature Med. 16, 998–1000 (2010)
Li, W. et al. Rapid induction and long-term self-renewal of primitive neural precursors from human embryonic stem cells by small molecule inhibitors. Proc. Natl Acad. Sci. USA 108, 8299–8304 (2011)
Krishnan, V. et al. Histone H4 lysine 16 hypoacetylation is associated with defective DNA repair and premature senescence in Zmpste24-deficient mice. Proc. Natl Acad. Sci. USA 108, 12325–12330 (2011)
Cheung, I. et al. Developmental regulation and individual differences of neuronal H3K4me3 epigenomes in the prefrontal cortex. Proc. Natl Acad. Sci. USA 107, 8824–8829 (2010)
Xie, W. et al. Proteasome inhibition modeling nigral neuron degeneration in Parkinson's disease. J. Neurochem. 115, 188–199 (2010)
Liu, G. H. et al. Targeted gene correction of laminopathy-associated LMNA mutations in patient-specific iPSCs. Cell Stem Cell 8, 688–694 (2011)
Suzuki, K. et al. Highly efficient transient gene expression and gene targeting in primate embryonic stem cells with helper-dependent adenoviral vectors. Proc. Natl Acad. Sci. USA 105, 13781–13786 (2008)
Li, M. et al. Efficient correction of hemoglobinopathy-causing mutations by homologous recombination in integration-free patient iPSCs. Cell Res. 21, 1740–1744 (2011)
Aizawa, E. et al. Efficient and accurate homologous recombination in hESCs and hiPSCs using helper-dependent adenoviral vectors. Mol. Ther. 20, 424–431 (2012)
Rudenko, I. N., Chia, R. & Cookson, M. R. Is inhibition of kinase activity the only therapeutic strategy for LRRK2-associated Parkinson's disease? BMC Med. 10, 20 (2012)
Kanao, T. et al. Activation of FoxO by LRRK2 induces expression of proapoptotic proteins and alters survival of postmitotic dopaminergic neuron in Drosophila . Hum. Mol. Genet. 19, 3747–3758 (2010)
Gehrke, S., Imai, Y., Sokol, N. & Lu, B. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature 466, 637–641 (2010)
Nichols, R. J. et al. 14-3-3 binding to LRRK2 is disrupted by multiple Parkinson's disease-associated mutations and regulates cytoplasmic localization. Biochem. J. 430, 393–404 (2010)
Dzamko, N. et al. Inhibition of LRRK2 kinase activity leads to dephosphorylation of Ser(910)/Ser(935), disruption of 14–3-3 binding and altered cytoplasmic localization. Biochem. J. 430, 405–413 (2010)
Scaffidi, P. & Misteli, T. Lamin A-dependent nuclear defects in human aging. Science 312, 1059–1063 (2006)
Poulopoulos, M. et al. Clinical and Pathological Characteristics of LRRK2 G2019S Patients with PD. J. Mol. Neurosci. 47, 139–143 (2012)
Thaler, A., Mirelman, A., Gurevich, T., Simon, E., Orr-Urtreger, A., Marder, K., Bressman, S. & Giladi, N. Lower cognitive performance in healthy G2019S LRRK2 mutation carriers. Neurology 79, 1027–1032 (2012)
Tiscornia, G., Vivas, E. L. & Belmonte, J. C. Diseases in a dish: modeling human genetic disorders using induced pluripotent cells. Nature Med. 17, 1570–1576 (2011)
Acknowledgements
We would like to thank K. Mitani, P. Ng, A. Lieber, Y. Imai, M. A. Miyawaki, Filocamo, S. Goldwurm, Telethon Genetic Biobank Network for providing constructs and cells (the fibroblast samples were obtained from the “Cell Line and DNA Biobank from patients affected by Genetic Diseases” (G. Gaslini Institute)-Telethon Genetic Biobank Network (project no. GTB07001)); Neurological Tissue Bank of the Biobank-Hospital Clínic-IDIBAPS for providing human brain tissue; F. Gage, M. Hetzer, J. Yao, Y. Mu, D. Yu, E. Gelpí, X. M. Wang, X. Wang, G. Bai and Z. J. Liu for helpful discussions; M. Joens and J. Fitzpatrick of the Waitt Advanced Biophotonics Core Facility for performing TEM analysis; M. Marti for imaging, teratoma and karyotyping analysis; F. Osakada for statistics analysis; and M. Schwarz, P. Schwarz and L. Laricchia-Robbio for administrative help. G.-H.L. is supported by the Thousand Young Talents program of China, the National Laboratory of Biomacromolecules, the Strategic Priority Research Program of the Chinese Academy of Sciences, the National Natural Science Foundation of China (NSFC) (81271266 and 31222039), and the Beijing Municipal Natural Science Foundation. J.Q. was partly supported by an AFAR/Ellison Medical Foundation postdoctoral fellowship. K.S. was partly supported by a Uehara Memorial Foundation research fellowship. E.N. was partly supported by an F.M. Kirby Foundation postdoctoral fellowship. X.X. is supported by NSFC (31201111). B.R. was supported by a US National Institute of Health (NIH) grant (ES017166) and the Ludwig Institute for Cancer Research. J.Y. was supported by an NIH grant (P41 RR011823). J.C.I.B. was supported by grants from the Glenn Foundation, G. Harold and Leila Y. Mathers Charitable Foundation, Sanofi, the California Institute of Regenerative Medicine, the Ellison Medical Foundation, the Helmsley Charitable Trust, ERA-Net Neuron, MINECO and Fundacion Cellex.
Author information
Authors and Affiliations
Contributions
G.-H.L., J.Q., K.S. prepared the figures, designed and performed all in vitro experiments. E.N. and N.M. designed and performed in vivo experiments. A.G., J.K., R.D.S., X.X., W.Z., Y.L., S.R. and C.R.E. provided technical assistance. I.D. performed teratoma studies. F.Y. generated microarray data. M.L. performed FISH and DNA methylation assays. B.R., U.W. and A.K. performed and analysed epigenetic studies. J.T. and J.Y.III performed proteomic studies. G.-H.L., J.Q., K.S., E.N., I.S.-M. and J.C.I.B. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary Figures 1-18 and Supplementary references. (PDF 3162 kb)
Supplementary Data
This file contains Supplementary Tables 1-5. (XLS 135 kb)
In-1 mediated restoration of cellular morphology
The In-1 mediated restoration of cellular morphology in late passage LRRK2 G2019S NSCs. 5 mM In-1 was added to passage 18 ipsNSCs-LK2(GS/GS), and then the cells were cultured for 5 days. Cells were imaged every 10 min for the 5 day duration. (MOV 18568 kb)
Rights and permissions
About this article
Cite this article
Liu, GH., Qu, J., Suzuki, K. et al. Progressive degeneration of human neural stem cells caused by pathogenic LRRK2. Nature 491, 603–607 (2012). https://doi.org/10.1038/nature11557
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11557
This article is cited by
-
Rapid and high-purity differentiation of human medium spiny neurons reveals LMNB1 hypofunction and subtype necessity in modeling Huntington’s disease
Inflammation and Regeneration (2024)
-
Promising biomarkers and therapeutic targets for the management of Parkinson's disease: recent advancements and contemporary research
Metabolic Brain Disease (2023)
-
Parkinson-causing mutations in LRRK2 impair the physiological tetramerization of endogenous α-synuclein in human neurons
npj Parkinson's Disease (2022)
-
Genomic, transcriptomic, and metabolomic profiles of hiPSC-derived dopamine neurons from clinically discordant brothers with identical PRKN deletions
npj Parkinson's Disease (2022)
-
hPSC gene editing for cardiac disease therapy
Pflügers Archiv - European Journal of Physiology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.