Abstract
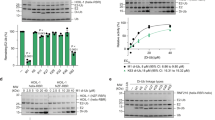
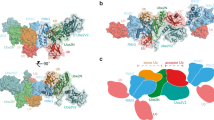
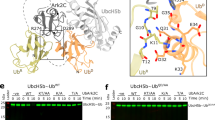
Ubiquitin modification is mediated by a large family of specificity determining ubiquitin E3 ligases. To facilitate ubiquitin transfer, RING E3 ligases bind both substrate and a ubiquitin E2 conjugating enzyme linked to ubiquitin via a thioester bond, but the mechanism of transfer has remained elusive. Here we report the crystal structure of the dimeric RING domain of rat RNF4 in complex with E2 (UbcH5A) linked by an isopeptide bond to ubiquitin. While the E2 contacts a single protomer of the RING, ubiquitin is folded back onto the E2 by contacts from both RING protomers. The carboxy-terminal tail of ubiquitin is locked into an active site groove on the E2 by an intricate network of interactions, resulting in changes at the E2 active site. This arrangement is primed for catalysis as it can deprotonate the incoming substrate lysine residue and stabilize the consequent tetrahedral transition-state intermediate.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kravtsova-Ivantsiv, Y. & Ciechanover, A. Non-canonical ubiquitin-based signals for proteasomal degradation. J. Cell Sci. 125, 539–548 (2012)
Budhidarmo, R., Nakatani, Y. & Day, C. L. RINGs hold the key to ubiquitin transfer. Trends Biochem. Sci. 37, 58–65 (2012)
Plechanovová, A. et al. Mechanism of ubiquitylation by dimeric RING ligase RNF4. Nature Struct. Mol. Biol. 18, 1052–1059 (2011)
Galanty, Y., Belotserkovskaya, R., Coates, J. & Jackson, S. P. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev. 26, 1179–1195 (2012)
Luo, K., Zhang, H., Wang, L., Yuan, J. & Lou, Z. Sumoylation of MDC1 is important for proper DNA damage response. EMBO J. 31, 3008–3019 (2012)
Yin, Y. et al. SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev. 26, 1196–1208 (2012)
Lallemand-Breitenbach, V. et al. Arsenic degrades PML or PML-RARα through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nature Cell Biol. 10, 547–555 (2008)
Tatham, M. H. et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nature Cell Biol. 10, 538–546 (2008)
Liew, C. W., Sun, H., Hunter, T. & Day, C. L. RING domain dimerization is essential for RNF4 function. Biochem. J. 431, 23–29 (2010)
Mace, P. D. et al. Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment. J. Biol. Chem. 283, 31633–31640 (2008)
Bentley, M. L. et al. Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. EMBO J. 30, 3285–3297 (2011)
Bosanac, I. et al. Modulation of K11-linkage formation by variable loop residues within UbcH5A. J. Mol. Biol. 408, 420–431 (2011)
Bosanac, I. et al. Ubiquitin binding to A20 ZnF4 is required for modulation of NF-kappaB signaling. Mol. Cell 40, 548–557 (2010)
Zhang, L. et al. The IDOL-UBE2D complex mediates sterol-dependent degradation of the LDL receptor. Genes Dev. 25, 1262–1274 (2011)
Hamilton, K. S. et al. Structure of a conjugating enzyme-ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail. Structure 9, 897–904 (2001)
Pruneda, J. N., Stoll, K. E., Bolton, L. J., Brzovic, P. S. & Klevit, R. E. Ubiquitin in motion: structural studies of the ubiquitin-conjugating enzyme∼ubiquitin conjugate. Biochemistry 50, 1624–1633 (2011)
Wickliffe, K. E., Lorenz, S., Wemmer, D. E., Kuriyan, J. & Rape, M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell 144, 769–781 (2011)
Sakata, E. et al. Crystal structure of UbcH5b∼ubiquitin intermediate: insight into the formation of the self-assembled E2∼Ub conjugates. Structure 18, 138–147 (2010)
Kamadurai, H. B. et al. Insights into ubiquitin transfer cascades from a structure of a UbcH5B∼ubiquitin-HECT(NEDD4L) complex. Mol. Cell 36, 1095–1102 (2009)
Wu, P. Y. et al. A conserved catalytic residue in the ubiquitin-conjugating enzyme family. EMBO J. 22, 5241–5250 (2003)
Yunus, A. A. & Lima, C. D. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nature Struct. Mol. Biol. 13, 491–499 (2006)
Yunus, A. A. & Lima, C. D. Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol. Cell 35, 669–682 (2009)
Reverter, D. & Lima, C. D. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature 435, 687–692 (2005)
Saha, A., Lewis, S., Kleiger, G., Kuhlman, B. & Deshaies, R. J. Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol. Cell 42, 75–83 (2011)
Gareau, J. R., Reverter, D. & Lima, C. D. Determinants of small ubiquitin-like modifier 1 (SUMO1) protein specificity, E3 ligase, and SUMO-RanGAP1 binding activities of nucleoporin RanBP2. J. Biol. Chem. 287, 4740–4751 (2012)
Calabrese, M. F. et al. A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases. Nature Struct. Mol. Biol. 18, 947–949 (2011)
Schreiber, A. et al. Structural basis for the subunit assembly of the anaphase-promoting complex. Nature 470, 227–232 (2011)
Brzovic, P. S., Lissounov, A., Christensen, D. E., Hoyt, D. W. & Klevit, R. E. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol. Cell 21, 873–880 (2006)
Martin, S. F., Hattersley, N., Samuel, I. D., Hay, R. T. & Tatham, M. H. A fluorescence-resonance-energy-transfer-based protease activity assay and its use to monitor paralog-specific small ubiquitin-like modifier processing. Anal. Biochem. 363, 83–90 (2007)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007)
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011)
Vijay-Kumar, S., Bugg, C. E. & Cook, W. J. Structure of ubiquitin refined at 1.8 A resolution. J. Mol. Biol. 194, 531–544 (1987)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007)
Lebedev, A. A. et al. JLigand: a graphical tool for the CCP4 template-restraint library. Acta Crystallogr. D 68, 431–440 (2012)
Zhang, M. et al. Chaperoned ubiquitylation—crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol. Cell 20, 525–538 (2005)
Shevchenko, A., Tomas, H., Havlis, J., Olsen, J. V. & Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nature Protocols 1, 2856–2860 (2006)
Nielsen, M. L. et al. Iodoacetamide-induced artifact mimics ubiquitination in mass spectrometry. Nature Methods 5, 459–460 (2008)
Cox, J. et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 (2011)
Acknowledgements
We thank M. Alphey and E. Branigan for assistance with data collection. His–UBE1 was a gift from the Division of Signal Transduction Therapy, University of Dundee. CHIP was a gift from A. Knebel and P. Cohen. A.P. was funded by the Wellcome Trust. This work was supported by a grant to R.T.H. from Cancer Research UK. Structural biology was supported by Scottish Funding Council (ref SULSA) and Wellcome Trust (program grant JHN).
Author information
Authors and Affiliations
Contributions
A.P. cloned, expressed and purified proteins, carried out structural analysis, conducted biochemical experiments and interpreted the data. E.G.J. purified recombinant proteins and carried out biochemical analysis. M.H.T. carried out mass spectrometry analysis. J.H.N. contributed to structural analysis and data analysis. A.P., J.H.N. and R.T.H. wrote the paper. R.T.H. conceived the project and contributed to data analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Table 1, Supplementary Figures 1-17 and additional references. (PDF 4767 kb)
Rights and permissions
About this article
Cite this article
Plechanovová, A., Jaffray, E., Tatham, M. et al. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 489, 115–120 (2012). https://doi.org/10.1038/nature11376
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11376
This article is cited by
-
UBE2A and UBE2B are recruited by an atypical E3 ligase module in UBR4
Nature Structural & Molecular Biology (2024)
-
Crystal structure of rice APIP6 reveals a new dimerization mode of RING-type E3 ligases that facilities the construction of its working model
Phytopathology Research (2023)
-
Cryo-EM structures of Uba7 reveal the molecular basis for ISG15 activation and E1-E2 thioester transfer
Nature Communications (2023)
-
Efficient determination of the accessible conformation space of multi-domain complexes based on EPR PELDOR data
Journal of Biomolecular NMR (2023)
-
Structure of the nutrient-sensing hub GATOR2
Nature (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.