Abstract
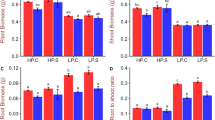
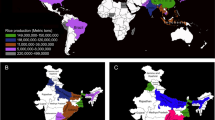
As an essential macroelement for all living cells, phosphorus is indispensable in agricultural production systems. Natural phosphorus reserves are limited1, and it is therefore important to develop phosphorus-efficient crops. A major quantitative trait locus for phosphorus-deficiency tolerance, Pup1, was identified in the traditional aus-type rice variety Kasalath about a decade ago2,3. However, its functional mechanism remained elusive4,5 until the locus was sequenced, showing the presence of a Pup1-specific protein kinase gene6, which we have named phosphorus-starvation tolerance 1 (PSTOL1). This gene is absent from the rice reference genome and other phosphorus-starvation-intolerant modern varieties7,8. Here we show that overexpression of PSTOL1 in such varieties significantly enhances grain yield in phosphorus-deficient soil. Further analyses show that PSTOL1 acts as an enhancer of early root growth, thereby enabling plants to acquire more phosphorus and other nutrients. The absence of PSTOL1 and other genes—for example, the submergence-tolerance gene SUB1A—from modern rice varieties underlines the importance of conserving and exploring traditional germplasm. Introgression of this quantitative trait locus into locally adapted rice varieties in Asia and Africa is expected to considerably enhance productivity under low phosphorus conditions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Data deposits
GenBank protein accession numbers for OsPupK04-1, OsPupK05-1, OsPupK20-2, OsPupK29-1 and PSTOL1/OsPupK46-2 are BAH79993, BAH79994, BAK26565, BAH80018 and BAK26566, respectively.
References
Van Kauwenbergh, S. J. World Phosphate Rock Reserves and Resources http://www.ifdc.org/getdoc/56358fb1-fc9b-49ba-92fe-187dc08e9586/T-75_World_Phosphate_Rock_Reserves_and_Resources (International Fertilizer Development Center, 2010)
Wissuwa, M., Yano, M. & Ae, N. Mapping of QTLs for phosphorus-deficiency tolerance in rice (Oryza sativa L.). Theor. Appl. Genet. 97, 777–783 (1998)
Wissuwa, M. & Ae, N. Genotypic variation for tolerance to phosphorus deficiency in rice and the potential for its exploitation in rice improvement. Plant Breed. 120, 43–48 (2001)
Wissuwa, M. Combining a modeling with a genetic approach in establishing associations between genetic and physiological effects in relation to phosphorus uptake. Plant Soil 269, 57–68 (2005)
Pariasca-Tanaka, J., Satoh, K., Rose, T., Mauleon, R. & Wissuwa, M. Stress response versus stress tolerance: a transcriptome analysis of two rice lines contrasting in tolerance to phosphorus deficiency. Rice 2, 167–185 (2009)
Heuer, S. et al. Comparative sequence analyses of the major quantitative trait locus Phosphorus uptake 1 (Pup1) reveal a complex genetic structure. Plant Biotechnol. J. 7, 456–471 (2009)
Chin, J. H. et al. Development and application of gene-based markers for the major rice QTL Phosphorus uptake 1. Theor. Appl. Genet. 120, 1073–1086 (2010)
Chin, J. H. et al. Developing rice with high yield under phosphorus deficiency: Pup1 sequence to application. Plant Physiol. 156, 1202–1216 (2011)
Haefele, S. M. & Hijmans, R. J. In Science, Technology, and Trade for Peace and Prosperity: Proceedings of the 26th International Rice Research Conference, New Delhi, India. (eds Aggarwal, P. K., Ladha, J. K., Singh, R. K., Devakumar, C. & Hardy, B. ) International Rice Research Institute, Indian Council of Agricultural Research, and National Academy of Agricultural Sciences, Los Baños, Philippines, and New Delhi, India. 297–308 (2007)
Haefele, S. M. & Hijmans, R. J. Soil quality in rainfed lowland rice. Rice Today 8, 30–31 (2009)
Dawe, D., Pandey, S. & Nelson, A. In Rice in the Global Economy. Strategic Research and Policy Issues for Food Security (eds Pandey, S. et al.) 5–35 (International Rice Research Institute, 2010)
Cordell, D., Drangert, J. O. & White, S. The story of phosphorus: global food security and food for thought. Glob. Environ. Change 19, 292–305 (2009)
Londo, J. P., Chiang, Y. C., Hung, K. H., Chiang, T. Y. & Schaal, B. A. Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc. Natl Acad. Sci. USA 103, 9578–9583 (2006)
Xu, K. et al. Sub1A is an ethylene-responsive-factor-like gene that confers submergence tolerance to rice. Nature 442, 705–708 (2006)
Manzanilla, D. O. et al. Submergence risks and farmers’ preferences: implications for breeding Sub1 rice in Southeast Asia. Agric. Syst. 104, 335–347 (2011)
Gowda, V. R. P. et al. Water uptake dynamics under progressive drought stress in diverse accessions of the OryzaSNP panel of rice (Oryza sativa). Func. Plant Bio. 39, 402–411 (2012)
Wissuwa, M., Wegner, J., Ae, N. & Yano, M. Substitution mapping of Pup1: a major QTL increasing phosphorus uptake of rice from a phosphorus-deficient soil. Theor. Appl. Genet. 105, 890–897 (2002)
Gregory, P. D., Barbari, S. & Hörz, W. Transcriptional control of phosphate-regulated genes. Food Technol. Biotechnol. 38, 295–303 (2000)
Yang, X. J. & Finnegan, P. M. Regulation of phosphate starvation responses in higher plants. Ann. Bot. (Lond.) 105, 513–526 (2010)
Dardick, C., Chen, J., Richter, T., Ouyang, S. & Ronald, P. The rice kinase database. A phylogenomic database for the rice kinome. Plant Physiol. 143, 579–586 (2007)
Vij, S., Giri, J., Dansana, P. K., Kapoor, S. & Tyagi, A. K. The receptor-like cytoplasmic kinase (OsRLCK) gene family in rice: organization, phylogenetic relationship, and expression during development and stress. Mol. Plant 1, 732–750 (2008)
Wang, X., Zafian, P., Choudhary, M. & Lawton, M. The PR5K receptor protein kinase from Arabidopsis thaliana is structurally related to a family of plant defense proteins. Proc. Natl Acad. Sci. USA 93, 2598–2602 (1996)
Bi, D., Cheng, Y. T., Li, X. & Zhang, Y. Activation of plant immune responses by a gain-of-function mutation in an atypical receptor-like kinase. Plant Physiol. 153, 1771–1779 (2010)
Bonardi, V. et al. Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature 437, 1179–1182 (2005)
Inukai, Y. et al. Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 17, 1387–1396 (2005)
Zhao, Y., Hu, Y., Dai, M., Huang, L. & Zhou, D.-X. The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell 21, 736–748 (2009)
Lohrmann, J. & Harter, K. Plant two-component signaling systems and the role of response regulators. Plant Physiol. 128, 363–369 (2002)
Scarpella, E., Simons, E. J. & Meijer, A. J. Multiple regulatory elements contribute to the vascular-specific expression of the rice HD-Zip gene Oshox1 in Arabidopsis. Plant Cell Physiol. 46, 1400–1410 (2005)
Kong, Z., Li, M., Yang, W., Xu, W. & Xue, Y. A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol. 141, 1376–1388 (2006)
Ouyang, J. et al. Identification and analysis of eight peptide transporter homologs in rice. Plant Sci. 179, 374–382 (2010)
Haldrup, A., Naver, H. & Scheller, H. V. The interaction between plastocyanin and photosystem I is inefficient in transgenic Arabidopsis plants lacking the PSI-N subunit of photosystem I. Plant J. 17, 689–698 (1999)
Pribil, M., Pesaresi, P., Hertle, A., Barbato, R. & Leister, D. Role of plastid protein phosphatase TAP38 in LHCII dephosphorylation and thylakoid electron flow. PLoS Biol. 8, e1000288 (2010)
Curtis, M. D. & Grossniklaus, U. A Gateway cloning vector set for high throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469 (2003)
Hiei, Y., Ishida, Y., Kasaoka, K. & Komari, T. Improved frequency of transformation in rice and maize by treatment of immature embryos with centrifugation and heat prior to infection with Agrobacterium tumefaciens. Plant Cell Tiss. Org. 87, 233–243 (2006)
Yoshida, S., Forno, D. A., Cock, J. H. & Gomez, K. A. Laboratory Manual for Physiological Studies of Rice 2nd edn 1–70 (International Rice Research Institute, 1972)
Jefferson, R. A., Kavanagh, T. A. & Bevan, M. W. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907 (1987)
Bernier, J., Kumar, A., Ramaiah, V., Spaner, D. & Atlin, G. A large-effect QTL for grain yield under reproductive-stage drought stress in upland rice. Crop Sci. 47, 507–518 (2007)
Bimpong, I. K. et al. Determination of genetic variability for physiological traits related to drought tolerance in African rice (Oryza glaberrima). J. Plant Breed. Crop. Sci. 3, 60–67 (2011)
Courtois, B. et al. Rice root architecture: meta-analysis from a drought QTL database. Rice 2, 115–128 (2009)
Gomez, M. S. et al. Mapping QTLs linked to physio-morphological and plant production traits under drought stress in rice (Oryza sativa L.) in the target environment. Am. J. Biochem. Biotechnol. 2, 161–169 (2006)
Lanceras, J. C., Pantuwan, G., Jongdee, B. & Toojinda, T. Quantitative trait loci associated with drought tolerance at reproductive stage in rice. Plant Physiol. 135, 384–399 (2004)
Ni, J. J., Wu, P., Senadhira, D. & Huang, N. Mapping QTLs for phosphorus deficiency tolerance in rice (Oryza sativa L.). Theor. Appl. Genet. 97, 1361–1369 (1998)
Thomson, M. J. et al. High-throughput single nucleotide polymorphism genotyping for breeding applications in rice using the BeadXpress platform. Mol. Breed. 29, 875–886 (2011)
Pritchard, J. K., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000)
Falush, D., Stephens, M. & Pritchard, J. K. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587 (2003)
Bradbury, P. J. et al. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635 (2007)
Miki, D. & Shimamoto, K. Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 45, 490–495 (2004)
Acknowledgements
We would like to thank A. Cruz, E. Ramos and L. Olivo for technical and secretarial support, the staff at the transformation laboratory, F. Rossi for help with the Pstol1 kinase assay, M. Akutsu for help with the analysis of transgenic plants, and S. Haefele and his team for their support. We thank J. Prasetiyono, M. Bustamam and S. Moeljopawiro for their long-term collaboration. This project has been primarily funded by the Generation Challenge Program (GCP) since 2005.
Author information
Authors and Affiliations
Contributions
R.G. cloned and transformed the PSTOL1 gene into IR64 and Nipponbare. R.G., J.P.T. and M.W. performed the phenotyping of transgenic plants. J.H.C. conducted the root meta-QTL analysis and J.H.C. and C.D. developed the IR64-Pup1 and IR74-Pup1 NILs. P.P. carried out the Pstol1 kinase assay. S.C. conducted the expression analysis of putative PSTOL1 downstream genes. E.M.T.M. provided advice about the experiments and I.S.-L. provided technical support and infrastructure for rice transformation. R.G., M.W. and S.H. designed the experiments and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures 1-9. (PDF 1264 kb)
Supplementary Data 1
This file contains Supplementary Table 1, which shows a summary of P starvation responsive genes. (XLS 224 kb)
Supplementary Data 2
This file contains Supplementary Table 2 showing a list of genes with constitutively altered expression in 35S::OsPSTOL1 roots. (XLS 40 kb)
Supplementary Data 3
This file contains Supplementary Table 3, which shows a list of primers used in this study. (XLS 26 kb)
Rights and permissions
About this article
Cite this article
Gamuyao, R., Chin, J., Pariasca-Tanaka, J. et al. The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature 488, 535–539 (2012). https://doi.org/10.1038/nature11346
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11346
This article is cited by
-
Fine mapping of the panicle length QTL qPL5 in rice
Molecular Breeding (2024)
-
Reviewing impacts of biotic and abiotic stresses on the regulation of phosphate homeostasis in plants
Journal of Plant Research (2024)
-
Involvement of reactive oxygen species in zinc-deficiency induced inhibition of crown root growth in maize plant
Plant and Soil (2024)
-
Rice straw-derived smoke water promotes rice root growth under phosphorus deficiency by modulating oxidative stress and photosynthetic gene expression
Scientific Reports (2023)
-
Super-pangenome analyses highlight genomic diversity and structural variation across wild and cultivated tomato species
Nature Genetics (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.