Abstract
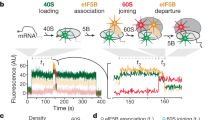
The initiation of translation establishes the reading frame for protein synthesis and is a key point of regulation1. Initiation involves factor-driven assembly at a start codon of a messenger RNA of an elongation-competent 70S ribosomal particle (in bacteria) from separated 30S and 50S subunits and initiator transfer RNA. Here we establish in Escherichia coli, using direct single-molecule tracking, the timing of initiator tRNA, initiation factor 2 (IF2; encoded by infB) and 50S subunit joining during initiation. Our results show multiple pathways to initiation, with orders of arrival of tRNA and IF2 dependent on factor concentration and composition. IF2 accelerates 50S subunit joining and stabilizes the assembled 70S complex. Transition to elongation is gated by the departure of IF2 after GTP hydrolysis, allowing efficient arrival of elongator tRNAs to the second codon presented in the aminoacyl-tRNA binding site (A site). These experiments highlight the power of single-molecule approaches to delineate mechanisms in complex multicomponent systems.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Laursen, B. S., Sorensen, H. P., Mortensen, K. K. & Sperling-Petersen, H. U. Initiation of protein synthesis in bacteria. Microbiol. Mol. Biol. Rev. 69, 101–123 (2005)
Kozak, M. Initiation of translation in prokaryotes and eukaryotes. Gene 234, 187–208 (1999)
Levene, M. J. et al. Zero-mode waveguides for single-molecule analysis at high concentrations. Science 299, 682–686 (2003)
Uemura, S. et al. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature 464, 1012–1017 (2010)
Milon, P. et al. The ribosome-bound initiation factor 2 recruits initiator tRNA to the 30S initiation complex. EMBO Rep. 11, 312–316 (2010)
Lockwood, A. H., Chakraborty, P. R. & Maitra, U. A complex between initiation factor IF2, guanosine triphosphate, and fMet-tRNA: an intermediate in initiation complex formation. Proc. Natl Acad. Sci. USA 68, 3122–3126 (1971)
Petersen, H. U., Roll, T., Grunberg-Manago, M. & Clark, B. F. Specific interaction of initiation factor IF2 of E. coli with formylmethionyl-tRNAfMet. Biochem. Biophys. Res. Commun. 91, 1068–1074 (1979)
Antoun, A., Pavlov, M. Y., Lovmar, M. & Ehrenberg, M. How initiation factors tune the rate of initiation of protein synthesis in bacteria. EMBO J. 25, 2539–2550 (2006)
Lockwood, A. H., Sarkar, P. & Maitra, U. Release of polypeptide chain initiation factor IF-2 during initiation complex formation. Proc. Natl Acad. Sci. USA 69, 3602–3605 (1972)
Caserta, E. et al. Translation initiation factor IF2 interacts with the 30 S ribosomal subunit via two separate binding sites. J. Mol. Biol. 362, 787–799 (2006)
Marshall, R. A., Aitken, C. E. & Puglisi, J. D. GTP hydrolysis by IF2 guides progression of the ribosome into elongation. Mol. Cell 35, 37–47 (2009)
Aitken, C. E. & Puglisi, J. D. Following the intersubunit conformation of the ribosome during translation in real time. Nature Struct. Mol. Biol. 17, 793–800 (2010)
Antoun, A., Pavlov, M. Y., Andersson, K., Tenson, T. & Ehrenberg, M. The roles of initiation factor 2 and guanosine triphosphate in initiation of protein synthesis. EMBO J. 22, 5593–5601 (2003)
Myasnikov, A. G. et al. Conformational transition of initiation factor 2 from the GTP- to GDP-bound state visualized on the ribosome. Nature Struct. Mol. Biol. 12, 1145–1149 (2005)
Blanchard, S. C., Kim, H. D., Gonzalez, R. L., Jr, Puglisi, J. D. & Chu, S. tRNA dynamics on the ribosome during translation. Proc. Natl Acad. Sci. USA 101, 12893–12898 (2004)
Marshall, R. A. Regulation of Protein Synthesis via Changes in Ribosome Conformation. PhD thesis, Stanford Univ. (2008)
Dorywalska, M. et al. Site-specific labeling of the ribosome for single-molecule spectroscopy. Nucleic Acids Res. 33, 182–189 (2005)
Blanchard, S. C., Gonzalez, R. L., Kim, H. D., Chu, S. & Puglisi, J. D. tRNA selection and kinetic proofreading in translation. Nature Struct. Mol. Biol. 11, 1008–1014 (2004)
Acknowledgements
Supported by National Institutes of Health grant GM51266 (J.D.P.) and the Japan Science and Technology Agency (S.U.).
Author information
Authors and Affiliations
Contributions
A.T., A.P. and S.U. conducted the experiments and performed the analysis; R.A.M. prepared and provided experimental materials; J.K. provided technical expertise with instrumentation and data processing; S.U. and J.D.P. designed experiments; and all authors discussed results and wrote the manuscript.
Corresponding authors
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-6, Supplementary Text and additional references. (PDF 1806 kb)
Rights and permissions
About this article
Cite this article
Tsai, A., Petrov, A., Marshall, R. et al. Heterogeneous pathways and timing of factor departure during translation initiation. Nature 487, 390–393 (2012). https://doi.org/10.1038/nature11172
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11172
This article is cited by
-
Real-time assembly of ribonucleoprotein complexes on nascent RNA transcripts
Nature Communications (2018)
-
AtaT blocks translation initiation by N-acetylation of the initiator tRNAfMet
Nature Chemical Biology (2017)
-
A conformational switch in initiation factor 2 controls the fidelity of translation initiation in bacteria
Nature Communications (2017)
-
Initiation of mRNA translation in bacteria: structural and dynamic aspects
Cellular and Molecular Life Sciences (2015)
-
Coordinated conformational and compositional dynamics drive ribosome translocation
Nature Structural & Molecular Biology (2013)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.