Abstract
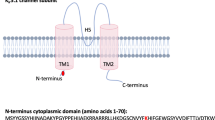
Hypertension affects one billion people and is a principal reversible risk factor for cardiovascular disease. Pseudohypoaldosteronism type II (PHAII), a rare Mendelian syndrome featuring hypertension, hyperkalaemia and metabolic acidosis, has revealed previously unrecognized physiology orchestrating the balance between renal salt reabsorption and K+ and H+ excretion1. Here we used exome sequencing to identify mutations in kelch-like 3 (KLHL3) or cullin 3 (CUL3) in PHAII patients from 41 unrelated families. KLHL3 mutations are either recessive or dominant, whereas CUL3 mutations are dominant and predominantly de novo. CUL3 and BTB-domain-containing kelch proteins such as KLHL3 are components of cullin–RING E3 ligase complexes that ubiquitinate substrates bound to kelch propeller domains2,3,4,5,6,7,8. Dominant KLHL3 mutations are clustered in short segments within the kelch propeller and BTB domains implicated in substrate9 and cullin5 binding, respectively. Diverse CUL3 mutations all result in skipping of exon 9, producing an in-frame deletion. Because dominant KLHL3 and CUL3 mutations both phenocopy recessive loss-of-function KLHL3 mutations, they may abrogate ubiquitination of KLHL3 substrates. Disease features are reversed by thiazide diuretics, which inhibit the Na–Cl cotransporter in the distal nephron of the kidney; KLHL3 and CUL3 are expressed in this location, suggesting a mechanistic link between KLHL3 and CUL3 mutations, increased Na–Cl reabsorption, and disease pathogenesis. These findings demonstrate the utility of exome sequencing in disease gene identification despite the combined complexities of locus heterogeneity, mixed models of transmission and frequent de novo mutation, and establish a fundamental role for KLHL3 and CUL3 in blood pressure, K+ and pH homeostasis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
Primary accessions
NCBI Reference Sequence
Data deposits
mRNA and protein sequences are available at NCBI under accession numbers NM_017415.2 and NP_059111.2 (KLHL3), NM_003590.3 and NP_003581.1 (CUL3); mutation data is available at dbSNP under batch accession 1056535.
References
Kahle, K. T., Ring, A. M. & Lifton, R. P. Molecular physiology of the WNK kinases. Annu. Rev. Physiol. 70, 329–355 (2008)
Lai, F. et al. Molecular characterization of KLHL3, a human homologue of the Drosophila kelch gene. Genomics 66, 65–75 (2000)
Furukawa, M., He, Y. J., Borchers, C. & Xiong, Y. Targeting of protein ubiquitination by BTB–Cullin 3–Roc1 ubiquitin ligases. Nature Cell Biol. 5, 1001–1007 (2003)
Prag, S. & Adams, J. C. Molecular phylogeny of the kelch-repeat superfamily reveals an expansion of BTB/kelch proteins in animals. BMC Bioinformatics 4, 42 (2003)
Stogios, P. J., Downs, G. S., Jauhal, J. J., Nandra, S. K. & Prive, G. G. Sequence and structural analysis of BTB domain proteins. Genome Biol. 6, R82 (2005)
Hudson, A. M. & Cooley, L. Phylogenetic, structural and functional relationships between WD- and kelch-repeat proteins. Subcell. Biochem. 48, 6–19 (2008)
Zimmerman, E. S., Schulman, B. A. & Zheng, N. Structural assembly of cullin-RING ubiquitin ligase complexes. Curr. Opin. Struct. Biol. 20, 714–721 (2010)
Sarikas, A., Hartmann, T. & Pan, Z. Q. The cullin protein family. Genome Biol. 12, 220 (2011)
Lo, S. C., Li, X., Henzl, M. T., Beamer, L. J. & Hannink, M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J. 25, 3605–3617 (2006)
Lifton, R. P., Gharavi, A. G. & Geller, D. S. Molecular mechanisms of human hypertension. Cell 104, 545–556 (2001)
Wilson, F. H. et al. Human hypertension caused by mutations in WNK kinases. Science 293, 1107–1112 (2001)
Choi, M. et al. K+ channel mutations in adrenal aldosterone-producing adenomas and hereditary hypertension. Science 331, 768–772 (2011)
Wilson, F. H. et al. Molecular pathogenesis of inherited hypertension with hyperkalemia: the Na–Cl cotransporter is inhibited by wild-type but not mutant WNK4. Proc. Natl Acad. Sci. USA 100, 680–684 (2003)
Yang, C. L., Angell, J., Mitchell, R. & Ellison, D. H. WNK kinases regulate thiazide-sensitive Na-Cl cotransport. J. Clin. Invest. 111, 1039–1045 (2003)
Kahle, K. T. et al. WNK4 regulates the balance between renal NaCl reabsorption and K+ secretion. Nature Genet. 35, 372–376 (2003)
Lalioti, M. D. et al. Wnk4 controls blood pressure and potassium homeostasis via regulation of mass and activity of the distal convoluted tubule. Nature Genet. 38, 1124–1132 (2006)
Ring, A. M. et al. WNK4 regulates activity of the epithelial Na+ channel in vitro and in vivo . Proc. Natl Acad. Sci. USA 104, 4020–4024 (2007)
Fairbrother, W. G., Yeh, R. F., Sharp, P. A. & Burge, C. B. Predictive identification of exonic splicing enhancers in human genes. Science 297, 1007–1013 (2002)
Bachmann, S., Bostanjoglo, M., Schmitt, R. & Ellison, D. H. Sodium transport-related proteins in the mammalian distal nephron—distribution, ontogeny and functional aspects. Anat. Embryol. (Berl.) 200, 447–468 (1999)
Welling, P. A. & Ho, K. A comprehensive guide to the ROMK potassium channel: form and function in health and disease. Am. J. Physiol. Renal Physiol. 297, F849–F863 (2009)
Zheng, N. et al. Structure of the Cul1–Rbx1–Skp1–F boxSkp2 SCF ubiquitin ligase complex. Nature 416, 703–709 (2002)
Ko, B. et al. RasGRP1 stimulation enhances ubiquitination and endocytosis of the sodium–chloride cotransporter. Am. J. Physiol. Renal Physiol. 299, F300–F309 (2010)
Simon, D. B. et al. Genetic heterogeneity of Bartter’s syndrome revealed by mutations in the K+ channel, ROMK. Nature Genet. 14, 152–156 (1996)
Karet, F. E. et al. Mutations in the gene encoding B1 subunit of H+-ATPase cause renal tubular acidosis with sensorineural deafness. Nature Genet. 21, 84–90 (1999)
Leviel, F. et al. The Na+-dependent chloride-bicarbonate exchanger SLC4A8 mediates an electroneutral Na+ reabsorption process in the renal cortical collecting ducts of mice. J. Clin. Invest. 120, 1627–1635 (2010)
Tong, K. I. et al. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol. Cell. Biol. 27, 7511–7521 (2007)
Wang, Y. et al. MMDB: annotating protein sequences with Entrez’s 3D-structure database. Nucleic Acids Res. 35, D298–D300 (2007)
Zhang, M. Q. Statistical features of human exons and their flanking regions. Hum. Mol. Genet. 7, 919–932 (1998)
Voets, T. et al. TRPM6 forms the Mg2+ influx channel involved in intestinal and renal Mg2+ absorption. J. Biol. Chem. 279, 19–25 (2004)
Fushimi, K. et al. Cloning and expression of apical membrane water channel of rat kidney collecting tubule. Nature 361, 549–552 (1993)
Choi, M. et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc. Natl Acad. Sci. USA 106, 19096–19101 (2009)
Li, H., Ruan, J. & Durbin, R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18, 1851–1858 (2008)
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009)
Robinson, J. T. et al. Integrative genomics viewer. Nature Biotechnol. 29, 24–26 (2011)
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007)
Abecasis, G. R., Cherny, S. S., Cookson, W. O. & Cardon, L. R. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nature Genet. 30, 97–101 (2002)
Rozen, S. & Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386 (2000)
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007)
Wang, Y., Geer, L. Y., Chappey, C., Kans, J. A. & Bryant, S. H. Cn3D: sequence and structure views for Entrez. Trends Biochem. Sci. 25, 300–302 (2000)
Acknowledgements
We thank the PHAII subjects, their families, and the health care professionals whose participation made this study possible; S. Umlauf and the staff of the Yale Center for Genome Analysis; J. Santosuosso; H. Tirrell and the staff of Beckman Coulter Genomics; V. Klump, Y. Lu, U. Scholl and J. Zhou for providing reagents; W. Hill for artistic assistance with Fig. 1d; and E. Boyden, S. Boyden, L. Cooley and M. Hochstrasser for helpful discussions. This work was supported in part by the Leducq Transatlantic Network on Hypertension and grants from the National Institutes of Health (P30-DK079310 and UL1-RR024139).
Author information
Authors and Affiliations
Contributions
L.M.B., M.C., K.A.C. and R.P.L. designed experiments and analysed data. L.M.B., C.J.N.-W. and I.R.T. performed experiments. A.F., H.R.T., G.C., M.L., R.D.G., B.A.S., A.P., M.J.V., M.E.D.F., S.A.S., M.G., F.E.K., J.R.T., J.R.S., K.M.K.-N., C.C.P., S.K.A., M.L.W., I.D.D., S.B.D., A.B., J.J.F., C.W.B., T.E.H., R.D.N., H.T., T.R.P.C., M.P., D.B., M.S., P.V., J.W.F., M.R., F.T., H.Z.A.-S., J.R., A.G.G. and B.G. recruited PHAII subjects and families. R.B. and S.M.M. directed the information technology and DNA sequencing infrastructure. L.M.B. and R.P.L. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-7 with legends and Supplementary Tables 1-4 (PDF 9330 kb)
Rights and permissions
About this article
Cite this article
Boyden, L., Choi, M., Choate, K. et al. Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature 482, 98–102 (2012). https://doi.org/10.1038/nature10814
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10814
This article is cited by
-
Update in genetic and epigenetic causes of hypertension
Cellular and Molecular Life Sciences (2024)
-
Severity of Autism Spectrum Disorder Symptoms Associated with de novo Variants and Pregnancy-Induced Hypertension
Journal of Autism and Developmental Disorders (2024)
-
Insights into the diverse mechanisms and effects of variant CUL3-induced familial hyperkalemic hypertension
Cell Communication and Signaling (2023)
-
Targeting E3 ubiquitin ligases and their adaptors as a therapeutic strategy for metabolic diseases
Experimental & Molecular Medicine (2023)
-
Restoration of Cullin3 gene expression enhances the improved effects of sonic hedgehog signaling activation for hypertension and attenuates the dysfunction of vascular smooth muscle cells
BioMedical Engineering OnLine (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.