Abstract
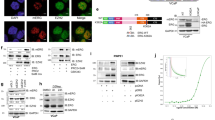
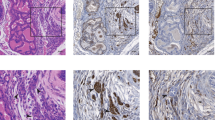
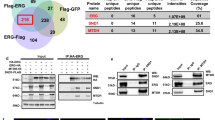
The proto-oncogenes ETV1, ETV4 and ETV5 encode transcription factors in the E26 transformation-specific (ETS) family, which includes the most frequently rearranged and overexpressed genes in prostate cancer1,2,3,4. Despite being critical regulators of development, little is known about their post-translational regulation. Here we identify the ubiquitin ligase COP1 (also known as RFWD2) as a tumour suppressor that negatively regulates ETV1, ETV4 and ETV5. ETV1, which is mutated in prostate cancer more often, was degraded after being ubiquitinated by COP1. Truncated ETV1 encoded by prostate cancer translocation TMPRSS2:ETV1 lacks the critical COP1 binding motifs and was 50-fold more stable than wild-type ETV1. Almost all patient translocations render ETV1 insensitive to COP1, implying that this confers a selective advantage to prostate epithelial cells. Indeed, COP1 deficiency in mouse prostate elevated ETV1 and produced increased cell proliferation, hyperplasia, and early prostate intraepithelial neoplasia. Combined loss of COP1 and PTEN enhanced the invasiveness of mouse prostate adenocarcinomas. Finally, rare human prostate cancer samples showed hemizygous loss of the COP1 gene, loss of COP1 protein, and elevated ETV1 protein while lacking a translocation event. These findings identify COP1 as a tumour suppressor whose downregulation promotes prostatic epithelial cell proliferation and tumorigenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Kumar-Sinha, C., Tomlins, S. A. & Chinnaiyan, A. M. Recurrent gene fusions in prostate cancer. Nature Rev. Cancer 8, 497–511 (2008)
Clark, J. P. & Cooper, C. S. ETS gene fusions in prostate cancer. Nature Rev. Urol. 6, 429–439 (2009)
Bartel, F. O., Higuchi, T. & Spyropoulos, D. D. Mouse models in the study of the Ets family of transcription factors. Oncogene 19, 6443–6454 (2000)
Gutierrez-Hartmann, A., Duval, D. L. & Bradford, A. P. ETS transcription factors in endocrine systems. Trends Endocrinol. Metab. 18, 150–158 (2007)
Wertz, I. E. et al. Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 303, 1371–1374 (2004)
Qi, L. et al. TRB3 links the E3 ubiquitin ligase COP1 to lipid metabolism. Science 312, 1763–1766 (2006)
Tomlins, S. A. et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature 448, 595–599 (2007)
Seo, H. S. et al. LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423, 995–999 (2003)
Bianchi, E. et al. Characterization of human constitutive photomorphogenesis protein 1, a RING finger ubiquitin ligase that interacts with Jun transcription factors and modulates their transcriptional activity. J. Biol. Chem. 278, 19682–19690 (2003)
Yi, C. & Deng, X. W. COP1 – from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 15, 618–625 (2005)
Pickart, C. M. Ubiquitin in chains. Trends Biochem. Sci. 25, 544–548 (2000)
Baert, J. L. et al. The E3 ubiquitin ligase complex component COP1 regulates PEA3 group member stability and transcriptional activity. Oncogene 29, 1810–1820 (2010)
Hermans, K. G. et al. Truncated ETV1, fused to novel tissue-specific genes, and full-length ETV1 in prostate cancer. Cancer Res. 68, 7541–7549 (2008)
Xin, L., Ide, H., Kim, Y., Dubey, P. & Witte, O. N. In vivo regeneration of murine prostate from dissociated cell populations of postnatal epithelia and urogenital sinus mesenchyme. Proc. Natl Acad. Sci. USA 100 (Suppl. 1). 11896–11903 (2003)
Leong, K. G., Wang, B. E., Johnson, L. & Gao, W. Q. Generation of a prostate from a single adult stem cell. Nature 456, 804–808 (2008)
Wu, X. et al. Generation of a prostate epithelial cell-specific Cre transgenic mouse model for tissue-specific gene ablation. Mech. Dev. 101, 61–69 (2001)
Shappell, S. B. et al. Prostate pathology of genetically engineered mice: definitions and classification. The consensus report from the Bar Harbor meeting of the Mouse Models of Human Cancer Consortium Prostate Pathology Committee. Cancer Res. 64, 2270–2305 (2004)
Ouyang, X. et al. Activator protein-1 transcription factors are associated with progression and recurrence of prostate cancer. Cancer Res. 68, 2132–2144 (2008)
Ali, I. U., Schriml, L. M. & Dean, M. Mutational spectra of PTEN/MMAC1 gene: a tumor suppressor with lipid phosphatase activity. J. Natl. Cancer Inst. 91, 1922–1932 (1999)
Yoshimoto, M. et al. Interphase FISH analysis of PTEN in histologic sections shows genomic deletions in 68% of primary prostate cancer and 23% of high-grade prostatic intra-epithelial neoplasias. Cancer Genet. Cytogenet. 169, 128–137 (2006)
King, J. C. et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nature Genet. 41, 524–526 (2009)
Carver, B. S. et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nature Genet. 41, 619–624 (2009)
Cai, C. et al. ETV1 is a novel androgen receptor-regulated gene that mediates prostate cancer cell invasion. Mol. Endocrinol. 21, 1835–1846 (2007)
de Launoit, Y. et al. The Ets transcription factors of the PEA3 group: transcriptional regulators in metastasis. Biochim. Biophys. Acta 1766, 79–87 (2006)
Whang, Y. E. et al. Inactivation of the tumor suppressor PTEN/MMAC1 in advanced human prostate cancer through loss of expression. Proc. Natl Acad. Sci. USA 95, 5246–5250 (1998)
Salghetti, S. E., Kim, S. Y. & Tansey, W. P. Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. EMBO J. 18, 717–726 (1999)
Bhatia, K. et al. Point mutations in the c-Myc transactivation domain are common in Burkitt’s lymphoma and mouse plasmacytomas. Nature Genet. 5, 56–61 (1993)
Albert, T., Urlbauer, B., Kohlhuber, F., Hammersen, B. & Eick, D. Ongoing mutations in the N-terminal domain of c-Myc affect transactivation in Burkitt’s lymphoma cell lines. Oncogene 9, 759–763 (1994)
Clark, H. M. et al. Mutations in the coding region of c-MYC in AIDS-associated and other aggressive lymphomas. Cancer Res. 54, 3383–3386 (1994)
Hayashi, S. & McMahon, A. P. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev. Biol. 244, 305–318 (2002)
Wang, S. et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell 4, 209–221 (2003)
Kayagaki, N. et al. DUBA: a deubiquitinase that regulates type I interferon production. Science 318, 1628–1632 (2007)
Shaulian, E., Zauberman, A., Ginsberg, D. & Oren, M. Identification of a minimal transforming domain of p53: negative dominance through abrogation of sequence-specific DNA binding. Mol. Cell. Biol. 12, 5581–5592 (1992)
Shevchenko, A., Tomas, H., Havlis, J., Olsen, J. V. & Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nature Protocols 1, 2856–2860 (2007)
Yi, E. C., Lee, H., Aebersold, R. & Goodlett, D. R. A microcapillary trap cartridge-microcapillary high-performance liquid chromatography electrospray ionization emitter device capable of peptide tandem mass spectrometry at the attomole level on an ion trap mass spectrometer with automated routine operation. Rapid Commun. Mass Spectrom. 17, 2093–2098 (2003)
Holm, M., Hardtke, C. S., Gaudet, R. & Deng, X. W. Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J. 20, 118–127 (2001)
Ang, L. H. et al. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1, 213–222 (1998)
Dentin, R. et al. Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature 449, 366–369 (2007)
Newton, K. et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell 134, 668–678 (2008)
Cunha, G. R. & Lung, B. The possible influence of temporal factors in androgenic responsiveness of urogenital tissue recombinants from wild-type and androgen-insensitive (Tfm) mice. J. Exp. Zool. 205, 181–193 (1978)
Jubb, A. M., Pham, T. Q., Frantz, G. D., Peale, F. V., Jr & Hillan, K. J. Quantitative in situ hybridization of tissue microarrays. Methods Mol. Biol. 326, 255–264 (2006)
Mehra, R. et al. Comprehensive assessment of TMPRSS2 and ETS family gene aberrations in clinically localized prostate cancer. Mod. Pathol. 20, 538–544 (2007)
O'Brien, C. et al. Functional genomics identifies ABCC3 as a mediator of taxane resistance in HER2-amplified breast cancer. Cancer Res. 68, 5380–5389 (2008)
Pandita, A., Aldape, K. D., Zadeh, G., Guha, A. & James, C. D. Contrasting in vivo and in vitro fates of glioblastoma cell subpopulations with amplified EGFR. Genes Chromosom. Cancer 39, 29–36 (2004)
Acknowledgements
We thank E. White and A. Chinnaiyan for reagents, B. Bolon for pathology support, and D. Dunlap for in situ hybridization.
Author information
Authors and Affiliations
Contributions
A.C.V. designed and performed in vitro experiments. A.C.V., K.G.L., C.Y. and W.-Q.G. designed and performed in vivo experiments. K.N. designed and generated the Cop1 mutant mice. R.V., R.F. and J.-A.H. generated monoclonal antibodies and performed immunohistochemistry. K.O. made constructs. J.L. performed bioinformatics analyses. S.M. and A.P. designed and performed FISH experiments. L.P. and D.A. designed and performed mass spectrometry experiments. S.S.C. and D.M.F. assessed the histopathology of mouse and human tissues. A.C.V., K.G.L., K.N., K.O., J.L., L.P., R.F., S.M., A.P., D.A., D.M.F. and V.M.D. prepared the manuscript and figures. A.C.V., K.G.L., K.N., C.Y., J.L., R.F., S.M., A.P., D.A., I.E.W., W.-Q.G., D.M.F. and V.M.D. contributed to the study design and data analyses.
Corresponding author
Ethics declarations
Competing interests
All authors were employees of Genentech, Inc.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-4 and Supplementary Figures 1-10 with legends. (PDF 24575 kb)
Rights and permissions
About this article
Cite this article
Vitari, A., Leong, K., Newton, K. et al. COP1 is a tumour suppressor that causes degradation of ETS transcription factors. Nature 474, 403–406 (2011). https://doi.org/10.1038/nature10005
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10005
This article is cited by
-
E26 transformation-specific transcription variant 5 in development and cancer: modification, regulation and function
Journal of Biomedical Science (2023)
-
CUL4B-DDB1-COP1-mediated UTX downregulation promotes colorectal cancer progression
Experimental Hematology & Oncology (2023)
-
HGF-mediated elevation of ETV1 facilitates hepatocellular carcinoma metastasis through upregulating PTK2 and c-MET
Journal of Experimental & Clinical Cancer Research (2022)
-
The E3 ligase COP1 promotes ERα signaling and suppresses EMT in breast cancer
Oncogene (2022)
-
COP1 Acts as a Ubiquitin Ligase for PCDH9 Ubiquitination and Degradation in Human Glioma
Molecular Neurobiology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.