Abstract
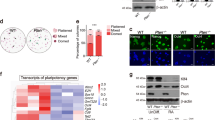
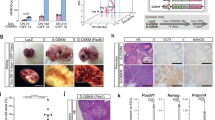
The rarity and inaccessibility of the earliest primordial germ cells (PGCs) in the mouse embryo thwart efforts to investigate molecular mechanisms of germ-cell specification. stella (also called Dppa3) marks the rare founder population of the germ lineage1,2. Here we differentiate mouse embryonic stem cells carrying a stella transgenic reporter into putative PGCs in vitro. The Stella+ cells possess a transcriptional profile similar to embryo-derived PGCs, and like their counterparts in vivo, lose imprints in a time-dependent manner. Using inhibitory RNAs to screen candidate genes for effects on the development of Stella+ cells in vitro, we discovered that Lin28, a negative regulator of let-7 microRNA processing3,4,5,6, is essential for proper PGC development. Furthermore, we show that Blimp1 (also called Prdm1), a let-7 target and a master regulator of PGC specification7,8,9, can rescue the effect of Lin28 deficiency during PGC development, thereby establishing a mechanism of action for Lin28 during PGC specification. Overexpression of Lin28 promotes formation of Stella+ cells in vitro and PGCs in chimaeric embryos, and is associated with human germ-cell tumours. The differentiation of putative PGCs from embryonic stem cells in vitro recapitulates the early stages of gamete development in vivo, and provides an accessible system for discovering novel genes involved in germ-cell development and malignancy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Payer, B. et al. Stella is a maternal effect gene required for normal early development in mice. Curr. Biol. 13, 2110–2117 (2003)
Payer, B. et al. Generation of stella-GFP transgenic mice: a novel tool to study germ cell development. Genesis 44, 75–83 (2006)
Heo, I. et al. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell 32, 276–284 (2008)
Newman, M. A., Thomson, J. M. & Hammond, S. M. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14, 1539–1549 (2008)
Rybak, A. et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nature Cell Biol. 10, 987–993 (2008)
Viswanathan, S. R., Daley, G. Q. & Gregory, R. I. Selective blockade of microRNA processing by Lin28. Science 320, 97–100 (2008)
Nie, K. et al. MicroRNA-mediated down-regulation of PRDM1/Blimp-1 in Hodgkin/Reed-Sternberg cells: a potential pathogenetic lesion in Hodgkin lymphomas. Am. J. Pathol. 173, 242–252 (2008)
Ohinata, Y. et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 436, 207–213 (2005)
Vincent, S. D. et al. The zinc finger transcriptional repressor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of primordial germ cells in the mouse. Development 132, 1315–1325 (2005)
Kurimoto, K. et al. Complex genome-wide transcription dynamics orchestrated by Blimp1 for the specification of the germ cell lineage in mice. Genes Dev. 22, 1617–1635 (2008)
Saitou, M. Specification of the germ cell lineage in mice. Front. Biosci. 14, 1068–1087 (2009)
Yamaji, M. et al. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nature Genet. 40, 1016–1022 (2008)
Geijsen, N. et al. Derivation of embryonic germ cells and male gametes from embryonic stem cells. Nature 427, 148–154 (2004)
Hayashi, K., Lopes, S. M., Tang, F. & Surani, M. A. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell 3, 391–401 (2008)
Wei, W. et al. Primordial germ cell specification from embryonic stem cells. PLoS ONE 3, e4013 (2008)
Reik, W. & Walter, J. Genomic imprinting: parental influence on the genome. Nature Rev. Genet. 2, 21–32 (2001)
Donovan, P. J. & de Miguel, M. P. Turning germ cells into stem cells. Curr. Opin. Genet. Dev. 13, 463–471 (2003)
Piskounova, E. et al. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J. Biol. Chem. 283, 21310–21314 (2008)
Chang, T. C. et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc. Natl Acad. Sci. USA 106, 3384–3389 (2009)
Dangi-Garimella, S. et al. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J. 28, 347–358 (2009)
Viswanathan, S. R. et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nature Genet. advance online publication 10.1038/ng.392 (31 May 2009)
Gidekel, S., Pizov, G., Bergman, Y. & Pikarsky, E. Oct-3/4 is a dose-dependent oncogenic fate determinant. Cancer Cell 4, 361–370 (2003)
Korkola, J. E. et al. Down-regulation of stem cell genes, including those in a 200-kb gene cluster at 12p13.31, is associated with in vivo differentiation of human male germ cell tumors. Cancer Res. 66, 820–827 (2006)
Cheng, L. Establishing a germ cell origin for metastatic tumors using OCT4 immunohistochemistry. Cancer 101, 2006–2010 (2004)
Hayashi, K. et al. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS ONE 3, e1738 (2008)
Ancelin, K. et al. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nature Cell Biol. 8, 623–630 (2006)
Garraway, L. A. et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 436, 117–122 (2005)
Shivdasani, R. A., Mayer, E. L. & Orkin, S. H. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature 373, 432–434 (1995)
Lu, J. et al. MicroRNA expression profiles classify human cancers. Nature 435, 834–838 (2005)
Keller, G. M. In vitro differentiation of embryonic stem cells. Curr. Opin. Cell Biol. 7, 862–869 (1995)
Kyba, M., Perlingeiro, R. C. & Daley, G. Q. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell 109, 29–37 (2002)
West, J. A., Park, I. H., Daley, G. Q. & Geijsen, N. In vitro generation of germ cells from murine embryonic stem cells. Nature Protocols 1, 2026–2036 (2006)
Beard, C., Hochedlinger, K., Plath, K., Wutz, A. & Jaenisch, R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis 44, 23–28 (2006)
Koshimizu, U., Watanabe, M. & Nakatsuji, N. Retinoic acid is a potent growth activator of mouse primordial germ cells in vitro . Dev. Biol. 168, 683–685 (1995)
Ginsburg, M., Snow, M. H. & McLaren, A. Primordial germ cells in the mouse embryo during gastrulation. Development 110, 521–528 (1990)
Stoger, R. et al. Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell 73, 61–71 (1993)
Yoon, B. J. et al. Regulation of DNA methylation of Rasgrf1 . Nature Genet. 30, 92–96 (2002)
Mager, J., Montgomery, N. D., de Villena, F. P. & Magnuson, T. Genome imprinting regulated by the mouse Polycomb group protein Eed. Nature Genet. 33, 502–507 (2003)
Lucifero, D., Mertineit, C., Clarke, H. J., Bestor, T. H. & Trasler, J. M. Methylation dynamics of imprinted genes in mouse germ cells. Genomics 79, 530–538 (2002)
Zaehres, H. & Daley, G. Q. Transgene expression and RNA interference in embryonic stem cells. Methods Enzymol. 420, 49–64 (2006)
Acknowledgements
We thank M. W. Lensch for comments on this manuscript; D. K. Gifford, G. Gerber, C. Reeder and J. Baughman for comments and input regarding microarray analysis; G. Losyev for flow cytometry expertise; and S. Winkler of the British Consulate for providing support for collaboration between the Daley and Surani laboratories. This study was supported by grants from the NIH, the NIH Director’s Pioneer Award of the NIH Roadmap for Medical Research, and by support from the germ cell program of the Harvard Stem Cell Institute. G.Q.D. is a recipient of the Burroughs Wellcome Fund Clinical Scientist Award in Translational Research.
Author Contributions J.A.W., project planning, experimental work, manuscript preparation; S.R.V., A.Y., A.T., K.C., I.-H.P., J.E.S., H.Z., A.P.-A., A.L.F., experimental work; M.A.S., contributed reagents and critical feedback; G.Q.D., project planning, data analysis and manuscript preparation.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-14 with Legends, Supplementary Tables 1-2 and Supplementary References. (PDF 1365 kb)
Rights and permissions
About this article
Cite this article
West, J., Viswanathan, S., Yabuuchi, A. et al. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature 460, 909–913 (2009). https://doi.org/10.1038/nature08210
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08210
This article is cited by
-
Application of Induced Pluripotent Stem Cells in Malignant Solid Tumors
Stem Cell Reviews and Reports (2023)
-
Lin28a maintains a subset of adult muscle stem cells in an embryonic-like state
Cell Research (2023)
-
Overexpression of Lin28A in neural progenitor cells in vivo does not lead to brain tumor formation but results in reduced spine density
Acta Neuropathologica Communications (2021)
-
Protein expression reveals a molecular sexual identity of avian primordial germ cells at pre-gonadal stages
Scientific Reports (2021)
-
Lin28, a major translation reprogramming factor, gains access to YB-1-packaged mRNA through its cold-shock domain
Communications Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.