Abstract
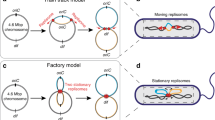
It has long been known that the 5′ to 3′ polarity of DNA synthesis results in both a leading and lagging strand at all replication forks1. Until now, however, there has been no evidence that leading or lagging strands are spatially organized in any way within a cell. Here we show that chromosome segregation in Escherichia coli is not random but is driven in a manner that results in the leading and lagging strands being addressed to particular cellular destinations. These destinations are consistent with the known patterns of chromosome segregation2,3. Our work demonstrates a new level of organization relating to the replication and segregation of the E. coli chromosome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
McInerney, P., Johnson, A., Katz, F. & O’Donnell, M. Characterization of a triple DNA polymerase replisome. Mol. Cell 27, 527–538 (2007)
Wang, X., Liu, X., Possoz, C. & Sherratt, D. J. The two Escherichia coli chromosome arms locate to separate cell halves. Genes Dev. 20, 1727–1731 (2006)
Nielsen, H. J. et al. The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Mol. Microbiol. 62, 331–338 (2006)
Thanbichler, M. & Shapiro, L. Getting organized—how bacterial cells move proteins and DNA. Nature Rev. Microbiol. 6, 28–40 (2008)
Wang, X., Possoz, C. & Sherratt, D. J. Dancing around the divisome: asymmetric chromosome segregation in Escherichia coli . Genes Dev. 19, 2367–2377 (2005)
Woldringh, C. L. & Nanninga, N. Structural and physical aspects of bacterial chromosome segregation. J. Struct. Biol. 156, 273–283 (2006)
Rocha, E. P. et al. A strand-specific model for chromosome segregation in bacteria. Mol. Microbiol. 49, 895–903 (2003)
Eykelenboom, J. K., Blackwood, J. K., Okely, E. & Leach, D. R. SbcCD causes a double-strand break at a DNA palindrome in the Escherichia coli chromosome. Mol. Cell 29, 644–651 (2008)
Pinder, D. J., Blake, C. E., Lindsey, J. C. & Leach, D. R. Replication strand preference for deletions associated with DNA palindromes. Mol. Microbiol. 28, 719–727 (1998)
Skarstad, K. & Boye, E. Degradation of individual chromosomes in recA mutants of Escherichia coli . J. Bacteriol. 175, 5505–5509 (1993)
Mason, D. J. & Powelson, D. M. Nuclear division as observed in live bacteria by a new technique. J. Bacteriol. 71, 474–479 (1956)
Khlebnikov, A. et al. Homogeneous expression of the P(BAD) promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology 147, 3241–3247 (2001)
Darmon, E. et al. SbcCD regulation and localization in Escherichia coli . J. Bacteriol. 189, 6686–6694 (2007)
Cairns, J. Mutation selection and the natural history of cancer. Nature 255, 197–200 (1975)
Rando, T. A. The immortal strand hypothesis: segregation and reconstruction. Cell 129, 1239–1243 (2007)
Kiel, M. J. et al. Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature 449, 238–242 (2007)
Waghmare, S. K. et al. Quantitative proliferation dynamics and random chromosome segregation of hair follicle stem cells. EMBO J. 27, 1309–1320 (2008)
Lansdorp, P. M. Immortal strands? Give me a break. Cell 129, 1244–1247 (2007)
Lew, D. J., Burke, D. J. & Dutta, A. The immortal strand hypothesis: how could it work? Cell 133, 21–23 (2008)
Niki, H., Yamaichi, Y. & Hiraga, S. Dynamic organization of chromosomal DNA in Escherichia coli . Genes Dev. 14, 212–223 (2000)
Yamaichi, Y. & Niki, H. migS, a cis-acting site that affects bipolar positioning of oriC on the Escherichia coli chromosome. EMBO J. 23, 221–233 (2004)
Thanbichler, M. & Shapiro, L. Chromosome organization and segregation in bacteria. J. Struct. Biol. 156, 292–303 (2006)
Danilova, O. et al. MukB colocalizes with the oriC region and is required for organization of the two Escherichia coli chromosome arms into separate cell halves. Mol. Microbiol. 65, 1485–1492 (2007)
Gitai, Z. et al. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120, 329–341 (2005)
Kruse, T. et al. Actin homolog MreB and RNA polymerase interact and are both required for chromosome segregation in Escherichia coli . Genes Dev. 20, 113–124 (2006)
Karczmarek, A. et al. DNA and origin region segregation are not affected by the transition from rod to sphere after inhibition of Escherichia coli MreB by A22. Mol. Microbiol. 65, 51–63 (2007)
Lemon, K. P. & Grossman, A. D. Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science 282, 1516–1519 (1998)
Rossi, M. L., Purohit, V., Brandt, P. D. & Bambara, R. A. Lagging strand replication proteins in genome stability and DNA repair. Chem. Rev. 106, 453–473 (2006)
Reyes-Lamothe, R., Possoz, C., Danilova, O. & Sherratt, D. J. Independent positioning and action of Escherichia coli replisomes in live cells. Cell 133, 90–102 (2008)
Possoz, C., Filipe, S. R., Grainge, I. & Sherratt, D. J. Tracking of controlled Escherichia coli replication fork stalling and restart at repressor-bound DNA in vivo . EMBO J. 25, 2596–2604 (2006)
Acknowledgements
We thank D. Sherratt for the gifts of plasmids pWX6, pLau43 and pLau44. We also thank E. Darmon and J. Blackwood for reading the manuscript. This work was supported by the Medical Research Council.
Author Contributions M.A.W. and D.R.F.L. conceived and designed the experiments; M.A.W. constructed all strains and plasmids apart from pDL1625, pDL1709 and pDL2542, which were constructed by J.K.E., E.W. and M.A.L.-V., respectively; M.A.W. performed the experiments; M.A.W. and D.R.F.L. analysed the data and wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Figure 1 with Legend and Supplementary Tables 1-3 (PDF 135 kb)
Rights and permissions
About this article
Cite this article
White, M., Eykelenboom, J., Lopez-Vernaza, M. et al. Non-random segregation of sister chromosomes in Escherichia coli. Nature 455, 1248–1250 (2008). https://doi.org/10.1038/nature07282
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature07282
This article is cited by
-
Nucleoid-mediated positioning and transport in bacteria
Current Genetics (2020)
-
Excision of Unstable Artificial Gene-Specific Inverted Repeats Mediates Scar-Free Gene Deletions in Escherichia coli
Applied Biochemistry and Biotechnology (2015)
-
RecA bundles mediate homology pairing between distant sisters during DNA break repair
Nature (2014)
-
Chromosome segregation by the Escherichia coli Min system
Molecular Systems Biology (2013)
-
Organization and segregation of bacterial chromosomes
Nature Reviews Genetics (2013)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.