Abstract
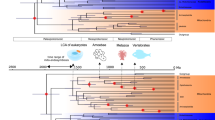
Biosignatures and structures in the geological record indicate that microbial life has inhabited Earth for the past 3.5 billion years or so1,2. Research in the physical sciences has been able to generate statements about the ancient environment that hosted this life3,4,5,6. These include the chemical compositions and temperatures of the early ocean and atmosphere. Only recently have the natural sciences been able to provide experimental results describing the environments of ancient life. Our previous work with resurrected proteins indicated that ancient life lived in a hot environment7,8. Here we expand the timescale of resurrected proteins to provide a palaeotemperature trend of the environments that hosted life from 3.5 to 0.5 billion years ago. The thermostability of more than 25 phylogenetically dispersed ancestral elongation factors suggest that the environment supporting ancient life cooled progressively by 30 °C during that period. Here we show that our results are robust to potential statistical bias associated with the posterior distribution of inferred character states, phylogenetic ambiguity, and uncertainties in the amino-acid equilibrium frequencies used by evolutionary models. Our results are further supported by a nearly identical cooling trend for the ancient ocean as inferred from the deposition of oxygen isotopes. The convergence of results from natural and physical sciences suggest that ancient life has continually adapted to changes in environmental temperatures throughout its evolutionary history.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Buick, R., Dunlop, J. S. R. & Groves, D. I. Stromatolite recognition in ancient rocks—an appraisal of irregularly laminated structures in an early Archean chert–barite unit from North Pole, Western Australia. Alcheringa 5, 161–181 (1981)
Hofmann, H. J., Grey, K., Hickman, A. H. & Thorpe, R. I. Origin of 3.45 Ga coniform stromatolites in Warrawoona Group, Western Australia. Geol. Soc. Am. Bull. 111, 1256–1262 (1999)
Knauth, L. P. & Lowe, D. R. Oxygen Isotope Geochemistry of Cherts from Onverwacht Group (3.4 billion years), Transvaal, South Africa, with implications for secular variations in isotopic composition of cherts. Earth Planet. Sci. Lett. 41, 209–222 (1978)
Knauth, L. P. & Lowe, D. R. High Archean climatic temperature inferred from oxygen isotope geochemistry of cherts in the 3.5 Ga Swaziland Supergroup, South Africa. Geol. Soc. Am. Bull. 115, 566–580 (2003)
Robert, F. & Chaussidon, M. A palaeotemperature curve for the Precambrian oceans based on silicon isotopes in cherts. Nature 443, 969–972 (2006)
Shen, Y., Buick, R. & Canfield, D. E. Isotopic evidence for microbial sulphate reduction in the early Archaean era. Nature 410, 77–81 (2001)
Gaucher, E. A. in Ancestral Sequence Reconstruction (ed. Liberles, D.A.) 20–33 (Oxford Univ. Press, Oxford, 2007)
Gaucher, E. A., Thomson, J. M., Burgan, M. F. & Benner, S. A. Inferring the palaeoenvironment of ancient bacteria on the basis of resurrected proteins. Nature 425, 285–288 (2003)
Liberles, D. A. Ancestral Sequence Reconstruction (Oxford Univ. Press, Oxford, 2007)
Williams, P. D., Pollock, D. D., Blackburne, B. P. & Goldstein, R. A. Assessing the accuracy of ancestral protein reconstruction methods. PLoS Comput. Biol. 2, e69 (2006)
Felsenstein, J. Cases in which parsimony or compatibility methods will be positively misleading. Syst. Zool. 27, 401–410 (1978)
Kelchner, S. A. & Thomas, M. A. Model use in phylogenetics: nine key questions. Trends Ecol. Evol. 22, 87–94 (2007)
Brooks, D. J. & Gaucher, E. A. in Ancestral Sequence Reconstruction (ed. Liberles, D.A.) 200–207 (Oxford Univ. Press, Oxford, 2007)
Cavalier-Smith, T. The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. Int. J. Syst. Evol. Microbiol. 52, 7–76 (2002)
Gromiha, M. M., Oobatake, M. & Sarai, A. Important amino acid properties for enhanced thermostability from mesophilic to thermophilic proteins. Biophys. Chem. 82, 51–67 (1999)
Battistuzzi, F. U., Feijao, A. & Hedges, S. B. A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol. Biol. 4, 44 (2004)
Ciccarelli, F. D. et al. Toward automatic reconstruction of a highly resolved tree of life. Science 311, 1283–1287 (2006)
Brochier, C. & Philippe, H. Phylogeny: a non-hyperthermophilic ancestor for Bacteria. Nature 417, 244 (2002)
Williams, R. A. D. & da Costa, M. S. in The Prokaryotes (eds Balows, A., Truper, H.G., Dworkin, M., Harder, W. & Schleifer, K.-H.) 3745–3753 (Springer, New York, 1992)
Hedges, S. B. et al. A genomic timescale for the origin of eukaryotes. BMC Evol. Biol. 1, 4 (2001)
Graur, D. & Martin, W. Reading the entrails of chickens: molecular timescales of evolution and the illusion of precision. Trends Genet. 20, 80–86 (2004)
Hoyle, F. History of Earth. Q. J. R. Astron. Soc. 13, 328–345 (1972)
Jaffres, J. B. D., Shields, G. A. & Wallmann, K. The oxygen isotope evolution of seawater: a critical review of a long-standing controversy and an improved geological water cycle model for the past 3.4 billion years. Earth Sci. Rev. 83, 83–122 (2007)
Kasting, J. F. et al. Paleoclimates, ocean depth, and the oxygen isotopic composition of seawater. Earth Planet. Sci. Lett. 252, 82–93 (2006)
Ward, D. M., Ferris, M. J., Nold, S. C. & Bateson, M. M. A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiol. Mol. Biol. Rev. 62, 1353–1370 (1998)
Altekar, G., Dwarkadas, S., Huelsenbeck, J. P. & Ronquist, F. Parallel metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics 20, 407–415 (2004)
Yang, Z. H. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13, 555–556 (1997)
Dillon, P. J. & Rosen, C. A. A rapid method for the construction of synthetic genes using the polymerase chain reaction. Biotechniques 9, 298–300 (1990)
Villalobos, A., Ness, J. E., Gustafsson, C., Minshull, J. & Govindarajan, S. Gene Designer: a synthetic biology tool for constructing artificial DNA segments. BMC Bioinformatics 7, 285 (2006)
Studier, F. W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005)
Acknowledgements
We thank A. Knoll for his comments on this research, and D. Brooks, F. Battistuzzi, R. Davis, K. Josephson, S. Sassi, R. Shaw, J. Szostak and S. Benner for their assistance. E.A.G. acknowledges support from the NASA Exobiology program. O.G. was supported by a NIH/NCRR grant to A. S. Edison and a National Science Foundation grant to S. J. Hagan. S.G. was supported financially by DNA2.0.
Author Contributions E.A.G. designed the study, performed the evolutionary analyses and circular dichroism experiments, analysed the results and wrote the manuscript. S.G. performed gene synthesis. O.G. performed circular dichroism experiments, fitted the data and analysed the results. All authors discussed the results and commented on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Figures
This file contains Supplementary Figures S1-S4 with Legends. (PDF 1326 kb)
Rights and permissions
About this article
Cite this article
Gaucher, E., Govindarajan, S. & Ganesh, O. Palaeotemperature trend for Precambrian life inferred from resurrected proteins. Nature 451, 704–707 (2008). https://doi.org/10.1038/nature06510
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature06510
This article is cited by
-
Evolution of optimal growth temperature in Asgard archaea inferred from the temperature dependence of GDP binding to EF-1A
Nature Communications (2024)
-
Comparative analysis of reconstructed ancestral proteins with their extant counterparts suggests primitive life had an alkaline habitat
Scientific Reports (2024)
-
A novel sequence-based predictor for identifying and characterizing thermophilic proteins using estimated propensity scores of dipeptides
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.