Abstract
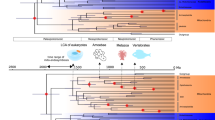
Determining the phylogenetic origin of mitochondria is key to understanding the ancestral mitochondrial symbiosis and its role in eukaryogenesis. However, the precise evolutionary relationship between mitochondria and their closest bacterial relatives remains hotly debated. The reasons include pervasive phylogenetic artefacts as well as limited protein and taxon sampling. Here we developed a new model of protein evolution that accommodates both across-site and across-branch compositional heterogeneity. We applied this site-and-branch-heterogeneous model (MAM60 + GFmix) to a considerably expanded dataset that comprises 108 mitochondrial proteins of alphaproteobacterial origin, and novel metagenome-assembled genomes from microbial mats, microbialites and sediments. The MAM60 + GFmix model fits the data much better and agrees with analyses of compositionally homogenized datasets with conventional site-heterogenous models. The consilience of evidence thus suggests that mitochondria are sister to the Alphaproteobacteria to the exclusion of MarineProteo1 and Magnetococcia. We also show that the ancestral presence of the crista-developing mitochondrial contact site and cristae organizing system (a mitofilin-domain-containing Mic60 protein) in mitochondria and the Alphaproteobacteria only supports their close relationship.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Sequencing data are deposited in NCBI GenBank under the BioProjects PRJNA315555, PRJNA438773, PRJNA754110, PRJNA754380, PRJNA752523 and PRJNA703749. Novel alphaproteobacterial MAGs and protein files (unaligned, aligned, and aligned and trimmed) are available at https://doi.org/10.6084/m9.figshare.14355845. Datasets and phylogenetic trees inferred in this study are available at https://doi.org/10.17632/dnbdzmjjkp.1.
Code availability
The GFmix model software is available at: https://www.mathstat.dal.ca/~tsusko/software.html
References
Roger, A. J., Muñoz-Gómez, S. A. & Kamikawa, R. The origin and diversification of mitochondria. Curr. Biol. 27, R1177–R1192 (2017).
Stairs, C. W., Leger, M. M. & Roger, A. J. Diversity and origins of anaerobic metabolism in mitochondria and related organelles. Phil. Trans. R. Soc. B 370, 20140326 (2015).
Müller, M. et al. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol. Mol. Biol. Rev. 76, 444–495 (2012).
Lane, N. & Martin, W. The energetics of genome complexity. Nature 467, 929–934 (2010).
Cavalier-Smith, T. Predation and eukaryote cell origins: a coevolutionary perspective. Int. J. Biochem. Cell Biol. 41, 307–322 (2009).
Spang, A. et al. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521, 173–179 (2015).
Zaremba-Niedzwiedzka, K. et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541, 353–358 (2017).
Eme, L., Spang, A., Lombard, J., Stairs, C. W. & Ettema, T. J. G. Archaea and the origin of eukaryotes. Nat. Rev. Microbiol. 15, 711–723 (2017).
Gray, M. W. Mitochondrial evolution. Cold Spring Harb. Perspect. Biol. 4, a011403 (2012).
Gray, M. W. Mosaic nature of the mitochondrial proteome: implications for the origin and evolution of mitochondria. Proc. Natl Acad. Sci. USA 112, 10133–10138 (2015).
Martijn, J., Vosseberg, J., Guy, L., Offre, P. & Ettema, T. J. G. Deep mitochondrial origin outside the sampled alphaproteobacteria. Nature 557, 101–105 (2018).
Fan, L. et al. Phylogenetic analyses with systematic taxon sampling show that mitochondria branch within Alphaproteobacteria. Nat. Ecol. Evol. 4, 1213–1219 (2020).
Viale, A. M. & Arakaki, A. K. The chaperone connection to the origins of the eukaryotic organelles. FEBS Lett. 341, 146–151 (1994).
Andersson, S. G. E. et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396, 133–140 (1998).
Wu, M. et al. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2, E69 (2004).
Fitzpatrick, D. A., Creevey, C. J. & McInerney, J. O. Genome phylogenies indicate a meaningful Α-proteobacterial phylogeny and support a grouping of the mitochondria with the Rickettsiales. Mol. Biol. Evol. 23, 74–85 (2006).
Williams, K. P., Sobral, B. W. & Dickerman, A. W. A robust species tree for the alphaproteobacteria. J. Bacteriol. 189, 4578–4586 (2007).
Sassera, D. et al. Phylogenomic evidence for the presence of a flagellum and cbb3 oxidase in the free-living mitochondrial ancestor. Mol. Biol. Evol. 28, 3285–3296 (2011).
Wang, Z. & Wu, M. Phylogenomic reconstruction indicates mitochondrial ancestor was an energy parasite. PLoS ONE 9, e110685 (2014).
Wang, Z. & Wu, M. An integrated phylogenomic approach toward pinpointing the origin of mitochondria. Sci. Rep. 5, 7949 (2015).
Ball, S. G., Bhattacharya, D. & Weber, A. P. M. Pathogen to powerhouse. Science 351, 659–660 (2016).
Thrash, J. C. et al. Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade. Sci. Rep. 1, 13 (2011).
Georgiades, K., Madoui, M.-A., Le, P., Robert, C. & Raoult, D. Phylogenomic analysis of Odyssella thessalonicensis fortifies the common origin of Rickettsiales, Pelagibacter ubique and Reclimonas americana mitochondrion. PLoS ONE 6, e24857 (2011).
Abhishek, A., Bavishi, A., Bavishi, A. & Choudhary, M. Bacterial genome chimaerism and the origin of mitochondria. Can. J. Microbiol. 57, 49–61 (2011).
Thiergart, T., Landan, G., Schenk, M., Dagan, T. & Martin, W. F. An evolutionary network of genes present in the eukaryote common ancestor polls genomes on eukaryotic and mitochondrial origin. Genome Biol. Evol. 4, 466–485 (2012).
Gawryluk, R. M. R. Evolutionary biology: a new home for the powerhouse? Curr. Biol. 28, R798–R800 (2018).
Eme, L., Sharpe, S. C., Brown, M. W. & Roger, A. J. On the age of eukaryotes: evaluating evidence from fossils and molecular clocks. Cold Spring Harb. Perspect. Biol. 6, a016139 (2014).
Betts, H. C. et al. Integrated genomic and fossil evidence illuminates life’s early evolution and eukaryote origin. Nat. Ecol. Evol. 2, 1556–1562 (2018).
Muñoz-Gómez, S. A. et al. An updated phylogeny of the Alphaproteobacteria reveals that the parasitic Rickettsiales and Holosporales have independent origins. eLife 8, e42535 (2019).
Luo, H. Evolutionary origin of a streamlined marine bacterioplankton lineage. ISME J. 9, 1423–1433 (2015).
Foster, P. G. Modeling compositional heterogeneity. Syst. Biol. 53, 485–495 (2004).
Rodríguez-Ezpeleta, N. & Embley, T. M. The SAR11 group of alpha-proteobacteria is not related to the origin of mitochondria. PLoS ONE 7, e30520 (2012).
Anantharaman, K. et al. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat. Commun. 7, 13219 (2016).
Graham, E. D., Heidelberg, J. F. & Tully, B. J. Potential for primary productivity in a globally-distributed bacterial phototroph. ISME J. 12, 1861–1866 (2018).
Delmont, T. O. et al. Nitrogen-fixing populations of Planctomycetes and Proteobacteria are abundant in surface ocean metagenomes. Nat. Microbiol. 3, 804–813 (2018).
Mehrshad, M., Amoozegar, M. A., Ghai, R., Shahzadeh Fazeli, S. A. & Rodriguez-Valera, F. Genome reconstruction from metagenomic data sets reveals novel microbes in the brackish waters of the Caspian Sea. Appl. Environ. Microbiol. 82, 1599–1612 (2016).
Tully, B. J., Sachdeva, R., Graham, E. D. & Heidelberg, J. F. 290 metagenome-assembled genomes from the Mediterranean Sea: a resource for marine microbiology. PeerJ 5, e3558 (2017).
Tully, B. J., Graham, E. D. & Heidelberg, J. F. The reconstruction of 2,631 draft metagenome-assembled genomes from the global oceans. Sci. Data 5, 170203 (2018).
Parks, D. H. et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2, 1533–1542 (2017).
Parks, D. H. et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004 (2018).
Gaston, D., Susko, E. & Roger, A. J. A phylogenetic mixture model for the identification of functionally divergent protein residues. Bioinformatics 27, 2655–2663 (2011).
Susko, E., Lincker, L. & Roger, A. J. Accelerated estimation of frequency classes in site-heterogeneous profile mixture models. Mol. Biol. Evol. 35, 1266–1283 (2018).
Muñoz-Gómez, S. A. et al. Additional Supplementary Data for ‘Site-and-branch-heterogeneous analyses of an expanded dataset favor mitochondria as sister to known Alphaproteobacteria. Mendeley Data https://doi.org/10.17632/dnbdzmjjkp.1 (2021).
Viklund, J., Ettema, T. J. G. & Andersson, S. G. E. Independent genome reduction and phylogenetic reclassification of the oceanic SAR11 clade. Mol. Biol. Evol. 29, 599–615 (2012).
Blanquart, S. & Lartillot, N. A Bayesian compound stochastic process for modeling nonstationary and nonhomogeneous sequence evolution. Mol. Biol. Evol. 23, 2058–2071 (2006).
Blanquart, S. & Lartillot, N. A site- and time-heterogeneous model of amino acid replacement. Mol. Biol. Evol. 25, 842–858 (2008).
Ferla, M. P., Thrash, J. C., Giovannoni, S. J. & Patrick, W. M. New rRNA gene-based phylogenies of the Alphaproteobacteria provide perspective on major groups, mitochondrial ancestry and phylogenetic instability. PLoS ONE 8, e83383 (2013).
Smith, D. R. Updating our view of organelle genome nucleotide landscape. Front. Genet. 3, 175 (2012).
Muñoz-Gómez, S. A. et al. Ancient homology of the mitochondrial contact site and cristae organizing system points to an endosymbiotic origin of mitochondrial cristae. Curr. Biol. 25, 1489–1495 (2015).
Muñoz-Gómez, S. A., Wideman, J. G., Roger, A. J. & Slamovits, C. H. The origin of mitochondrial cristae from Alphaproteobacteria. Mol. Biol. Evol. 34, 943–956 (2017).
Gutiérrez-Preciado, A. et al. Functional shifts in microbial mats recapitulate early Earth metabolic transitions. Nat. Ecol. Evol. 2, 1700–1708 (2018).
Saghaï, A. et al. Comparative metagenomics unveils functions and genome features of microbialite-associated communities along a depth gradient. Environ. Microbiol. 18, 4990–5004 (2016).
Saghaï, A. et al. Metagenome-based diversity analyses suggest a significant contribution of non-cyanobacterial lineages to carbonate precipitation in modern microbialites. Front. Microbiol. 6, 797 (2015).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Wu, Y.-W., Simmons, B. A. & Singer, S. W. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32, 605–607 (2016).
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015).
Eren, A. M. et al. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3, e1319 (2015).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015).
Alneberg, J. et al. Binning metagenomic contigs by coverage and composition. Nat. Methods 11, 1144–1146 (2014).
Li, D., Liu, C.-M., Luo, R., Sadakane, K. & Lam, T.-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015).
Kang, D. D. et al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7, e7359 (2019).
Sieber, C. M. K. et al. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 3, 836–843 (2018).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl. Acids Res 25, 3389–3402 (1997).
Katoh, K., Kuma, K., Toh, H. & Miyata, T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518 (2005).
Criscuolo, A. & Gribaldo, S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 10, 210 (2010).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Kannan, S., Rogozin, I. B. & Koonin, E. V. MitoCOGs: clusters of orthologous genes from mitochondria and implications for the evolution of eukaryotes. BMC Evol. Biol. 14, 237 (2014).
Capella-Gutiérrez, S., Silla-Martínez, J. M. & Gabaldón, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A. & Jermiin, L. S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017).
Menardo, F. et al. Treemmer: a tool to reduce large phylogenetic datasets with minimal loss of diversity. BMC Bioinformatics 19, 164 (2018).
Ali, R. H., Bogusz, M. & Whelan, S. Identifying clusters of high confidence homologies in multiple sequence alignments. Mol. Biol. Evol. 36, 2340–2351 (2019).
de Vienne, D. M., Ollier, S. & Aguileta, G. Phylo-MCOA: a fast and efficient method to detect outlier genes and species in phylogenomics using multiple co-inertia analysis. Mol. Biol. Evol. 29, 1587–1598 (2012).
Vaidya, G., Lohman, D. J. & Meier, R. SequenceMatrix: concatenation software for the fast assembly of multi‐gene datasets with character set and codon information. Cladistics 27, 171–180 (2011).
Muñoz-Gómez, S. A. et al. Alignments for 108 mitochondrial proteins of alphaproteobacterial origin, and alphaproteobacterial MAGs from microbial mats, microbialites, and sediments. figshare https://doi.org/10.6084/m9.figshare.14355845.v2 (2021).
Wang, H.-C., Minh, B. Q., Susko, E. & Roger, A. J. Modeling site heterogeneity with posterior mean site frequency profiles accelerates accurate phylogenomic estimation. Syst. Biol. 67, 216–235 (2018).
Schrempf, D., Lartillot, N. & Szöllősi, G. Scalable empirical mixture models that account for across-site compositional heterogeneity. Mol. Biol. Evol. 37, 3616–3631 (2020).
Lartillot, N. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 21, 1095–1109 (2004).
Lartillot, N., Rodrigue, N., Stubbs, D. & Richer, J. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst. Biol. 62, 611–615 (2013).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018).
Susko, E. Tests for two trees using likelihood methods. Mol. Biol. Evol. 31, 1029–1039 (2014).
Shimodaira, H. & Hasegawa, M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16, 1114 (1999).
Markowski, E. A Comparison of Methods for Constructing Confidence Sets of Phylogenetic Trees Using Maximum Likelihood. MSc thesis, Dalhousie Univ. (2021).
Lee, M. D. GToTree: a user-friendly workflow for phylogenomics. Bioinformatics 35, 4162–4164 (2019).
Acknowledgements
S.A.M.-G. is supported by an EMBO Postdoctoral Fellowship (ALTF 21-2020). We thank B. Curtis (Dalhousie University) and D. Salas-Leiva (Dalhousie University) for assistance with scripts, W. Valencia (Harvard University) and C. Calderon (Rutgers University) for advice on Python and R, and A. Gutiérrez-Preciado (Université Paris-Saclay) for assistance with uploading data to NCBI GenBank. This work was supported by the Moore-Simons Project on the Origin of the Eukaryotic Cell, Simons Foundation grants 735923LPI (https://doi.org/10.46714/735923LPI) awarded to A.J.R. and GBMF9739 (https://doi.org/10.37807/GBMF9739) awarded to P.L.G., and Discovery Grants from the Natural Sciences and Engineering Research Council of Canada awarded to A.J.R., E.S. and C.H.S.
Author information
Authors and Affiliations
Contributions
S.A.M.-G.: conceptualization, methodology, validation, formal analysis, investigation, data curation, writing—original draft, writing—review and editing, visualization, project administration, funding acquisition. E.S.: methodology, software, writing—review and editing. K.W.: validation, data curation, writing—review and editing. L.E.: resources, writing—review and editing. C.H.S.: resources, supervision, writing—review and editing, funding acquisition. D.M.: resources, writing—review and editing, funding acquisition. P.L.-G.: resources, writing—review and editing, funding acquisition. A.J.R.: conceptualization, methodology, validation, resources, supervision, project administration, writing—review and editing, funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review information Nature Ecology & Evolution thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Euler diagram that shows the relationships between recent phylogenomic sets of proteins used to address the phylogenetic placement of mitochondria.
Datasets include those comprised of mitochondrion- and nucleus-encoded proteins in the studies Wang and Wu 20, Martijn et al. 11, and this study. Nucleus-encoded proteins are in green, mitochondrion-encoded proteins in red, and both nucleus- and mitochondrion-encoded proteins in blue. Gene/protein names mostly follow the human gene nomenclature.
Extended Data Fig. 2 Summary of features for novel MAGs that belong to the MarineProteo1 clade and the Rickettsiales.
Branches highlighted in red show taxa used for phylogenetic analyses in this study. The dashed rectangle points to the secondary higher G + C% content of the genera Anaplasma and Neorickettsia in the family Anaplasmataceae. The Magnetococcia is at the base of the tree as an outgroup.
Extended Data Fig. 3 Branch support variation for the placement of mitochondria outside of the Alphaproteobacteria throughout the progressive removal of compositionally heterogenous sites.
Branch support values are SH-aLRT and UFBoot2+NNI and the removal of compositionally heterogeneous sites was done according to the ɀ and χ2 metrics. Support for the branch that groups mitochondria with all alphaproteobacteria (but excludes MarineProteo1 and the Magnetococcia) is always maximal (i.e., 100% SH-aLRT /100% UFBoot2+NNI). (a) Nucleus-encoded protein dataset. (b) Mitochondrion-encoded protein M1 dataset. (c) Mitochondrion-encoded protein M2 dataset.
Extended Data Fig. 4 Branch support variation for the placement of mitochondria when derived and compositionally biased Rickettsiales are included throughout the progressive removal of compositionally heterogenous sites.
Branch support values are SH-aLRT and UFBoot2+NNI and the removal of compositionally heterogeneous sites was done according to the ɀ and χ2 metrics. (a) Alphaproteobacteria-sister topology. Support for the branch that groups mitochondria with all alphaproteobacteria (but excludes MarineProteo1 and the Magnetococcia) is always maximal (i.e., 100% SH-aLRT /100% UFBoot2+NNI). (b) Rickettsiales-sister topology.
Extended Data Fig. 5 Schematic tree topologies used for calculating likelihood values using the MAM60 + GFmix model.
(a) Tree topologies derived from analyses of the untreated dataset of mitochondrion-, and nucleus-encoded proteins. (b) Tree topologies derived from analyses of a compositionally homogenized dataset of mitochondrion-, and nucleus-encoded proteins. (c) Tree topologies derived from analyses of the untreated dataset of nucleus-encoded proteins. (d) Tree topologies derived from analyses of a compositionally homogenized dataset of nucleus-encoded proteins. (e) Tree topologies derived from analyses of the untreated dataset of mitochondrion-encoded proteins. (f) Tree topologies derived from analyses of a compositionally homogenized dataset of mitochondrion-encoded proteins. Datasets were compositionally homogenized by removing the 50% most compositionally heterogeneous sites according to the ɀ metric.
Extended Data Fig. 6 UPGMAs dendrograms for G A R P/F I M N K Y distances among the marker proteins of alphaproteobacterial origin in eukaryotes used in this study.
(a) Mitochondrion- and nucleus-encoded proteins. (b) Nucleus-encoded proteins. (c). Mitochondrion-encoded proteins. Nucleus-encoded proteins are in green, mitochondrion-encoded proteins in red, and both nucleus- and mitochondrion-encoded proteins in blue. Gene/protein names mostly follow the human gene nomenclature.
Extended Data Fig. 7 Phylogenetic distribution of the Mitofilin-domain containing Mic60 in the Proteobacteria.
The Mitofilin-domain containing Mic60, as defined by the Pfam pHMM Mitofilin PF09731, is phylogenetically restructured to the Alphaproteobacteria to the exclusion of MarineProteo1 clade and the Magnetococcia. This protein is also conspicuously absent in the Gamma- and Zetaproteobacteria.
Supplementary information
Rights and permissions
About this article
Cite this article
Muñoz-Gómez, S.A., Susko, E., Williamson, K. et al. Site-and-branch-heterogeneous analyses of an expanded dataset favour mitochondria as sister to known Alphaproteobacteria. Nat Ecol Evol 6, 253–262 (2022). https://doi.org/10.1038/s41559-021-01638-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-021-01638-2
This article is cited by
-
Expanded phylogeny of extremely halophilic archaea shows multiple independent adaptations to hypersaline environments
Nature Microbiology (2024)
-
Recent findings in methanotrophs: genetics, molecular ecology, and biopotential
Applied Microbiology and Biotechnology (2024)
-
Incongruence in the phylogenomics era
Nature Reviews Genetics (2023)
-
Ecophysiology and genomics of the brackish water adapted SAR11 subclade IIIa
The ISME Journal (2023)
-
The virome of the last eukaryotic common ancestor and eukaryogenesis
Nature Microbiology (2023)