Abstract
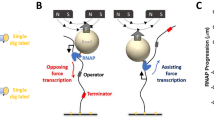
RNA polymerase II (RNAP II) is responsible for transcribing all messenger RNAs in eukaryotic cells during a highly regulated process that is conserved from yeast to human1, and that serves as a central control point for cellular function. Here we investigate the transcription dynamics of single RNAP II molecules from Saccharomyces cerevisiae against force and in the presence and absence of TFIIS, a transcription elongation factor known to increase transcription through nucleosomal barriers2. Using a single-molecule dual-trap optical-tweezers assay combined with a novel method to enrich for active complexes, we found that the response of RNAP II to a hindering force is entirely determined by enzyme backtracking3,4,5,6. Surprisingly, RNAP II molecules ceased to transcribe and were unable to recover from backtracks at a force of 7.5 ± 2 pN, only one-third of the force determined for Escherichia coli RNAP7,8. We show that backtrack pause durations follow a t-3/2 power law, implying that during backtracking RNAP II diffuses in discrete base-pair steps, and indicating that backtracks may account for most of RNAP II pauses. Significantly, addition of TFIIS rescued backtracked enzymes and allowed transcription to proceed up to a force of 16.9 ± 3.4 pN. Taken together, these results describe a regulatory mechanism of transcription elongation in eukaryotes by which transcription factors modify the mechanical performance of RNAP II, allowing it to operate against higher loads.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Roeder, R. G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21, 327–335 (1996)
Kireeva, M. L. et al. Nature of the nucleosomal barrier to RNA polymerase II. Mol. Cell 18, 97–108 (2005)
Komissarova, N. & Kashlev, M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc. Natl Acad. Sci. USA 94, 1755–1760 (1997)
Komissarova, N. & Kashlev, M. RNA polymerase switches between inactivated and activated states by translocating back and forth along the DNA and the RNA. J. Biol. Chem. 272, 15329–15338 (1997)
Nudler, E., Mustaev, A., Lukhtanov, E. & Goldfarb, A. The RNA–DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell 89, 33–41 (1997)
Shaevitz, J. W., Abbondanzieri, E. A., Landick, R. & Block, S. M. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature 426, 684–687 (2003)
Davenport, R. J., Wuite, G. J., Landick, R. & Bustamante, C. Single-molecule study of transcriptional pausing and arrest by E. coli RNA polymerase. Science 287, 2497–2500 (2000)
Wang, M. D. et al. Force and velocity measured for single molecules of RNA polymerase. Science 282, 902–907 (1998)
Hahn, S. Structure and mechanism of the RNA polymerase II transcription machinery. Nature Struct. Mol. Biol. 11, 394–403 (2004)
Komissarova, N., Kireeva, M. L., Becker, J., Sidorenkov, I. & Kashlev, M. Engineering of elongation complexes of bacterial and yeast RNA polymerases. Methods Enzymol. 371, 233–251 (2003)
Abbondanzieri, E. A., Greenleaf, W. J., Shaevitz, J. W., Landick, R. & Block, S. M. Direct observation of base-pair stepping by RNA polymerase. Nature 438, 460–465 (2005)
Moffitt, J., Chemla, Y., Izhaky, D. & Bustamante, C. Differential detection of dual traps improves the spatial resolution of optical tweezers. Proc. Natl Acad. Sci. USA 103, 9006–9011 (2006)
Bustamante, C., Marko, J. F., Siggia, E. D. & Smith, S. Entropic elasticity of lambda-phage DNA. Science 265, 1599–1600 (1994)
Neuman, K. C., Abbondanzieri, E. A., Landick, R., Gelles, J. & Block, S. M. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell 115, 437–447 (2003)
Izban, M. G. & Luse, D. S. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J. Biol. Chem. 267, 13647–13655 (1992)
Forde, N. R., Izhaky, D., Woodcock, G. R., Wuite, G. J. L. & Bustamante, C. Using mechanical force to probe the mechanism of pausing and arrest during continuous elongation by Escherichia coli RNA polymerase. Proc. Natl Acad. Sci. USA 99, 11682–11687 (2002)
Yin, H. et al. Transcription against an applied force. Science 270, 1653–1657 (1995)
Kulish, D. & Struhl, K. TFIIS enhances transcriptional elongation through an artificial arrest site in vivo. Mol. Cell. Biol. 21, 4162–4168 (2001)
Li, P. T. X., Collin, D., Smith, S. B., Bustamante, C. & Tinoco, I. Probing the mechanical folding kinetics of TAR RNA by hopping, force-jump, and force-ramp methods. Biophys. J. 90, 250–260 (2006)
Fish, R. N. & Kane, C. M. Promoting elongation with transcript cleavage stimulatory factors. Biochim. Biophys. Acta 1577, 287–307 (2002)
Borukhov, S., Lee, J. & Laptenko, O. Bacterial transcription elongation factors: new insights into molecular mechanism of action. Mol. Microbiol. 55, 1315–1324 (2005)
Jeon, C. & Agarwal, K. Fidelity of RNA polymerase II transcription controlled by elongation factor TFIIS. Proc. Natl Acad. Sci. USA 93, 13677–13682 (1996)
Awrey, D. E. et al. Yeast transcript elongation factor (TFIIS), structure and function. II: RNA polymerase binding, transcript cleavage, and read-through. J. Biol. Chem. 273, 22595–22605 (1998)
Weilbaecher, R. G., Awrey, D. E., Edwards, A. M. & Kane, C. M. Intrinsic transcript cleavage in yeast RNA polymerase II elongation complexes. J. Biol. Chem. 278, 24189–24199 (2003)
Reines, D. & Mote, J. Elongation factor SII-dependent transcription by RNA polymerase II through a sequence-specific DNA-binding protein. Proc. Natl Acad. Sci. USA 90, 1917–1921 (1993)
Palangat, M., Renner, D. B., Price, D. H. & Landick, R. A negative elongation factor for human RNA polymerase II inhibits the anti-arrest transcript-cleavage factor TFIIS. Proc. Natl Acad. Sci. USA 102, 15036–15041 (2005)
Kireeva, M. L., Lubkowska, L., Komissarova, N. & Kashlev, M. Assays and affinity purification of biotinylated and nonbiotinylated forms of double-tagged core RNA polymerase II from Saccharomyces cerevisiae. Methods Enzymol. 370, 138–155 (2003)
Smith, S. B., Cui, Y. & Bustamante, C. Optical-trap force transducer that operates by direct measurement of light momentum. Methods Enzymol. 361, 134–162 (2003)
Acknowledgements
We thank Y. R. Chemla, W. Cheng, M. Cruse, S. Dumont, N. R. Forde, B. Ibarra, D. Izhaky, C. Kane, S. Kostek, J. Moffit, J. M. R. Parrondo, M. Peris, S. Plyasunov, A. Ruiz and S. B. Smith for experimental assistance and helpful discussions. This research was supported by a DOE grant. E.A.G. was supported by a Jane Coffin Childs Postdoctoral Fellowship. S.W.G. was supported first by an EMBO Long Term Fellowship and subsequently by a Helen Hay Whitney Postdoctoral Fellowship. We dedicate this manuscript to Jason Choy.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures S1- S8, Supplementary Table S1, Supplementary Methods, Supplementary Equations and additional references (PDF 600 kb)
Rights and permissions
About this article
Cite this article
Galburt, E., Grill, S., Wiedmann, A. et al. Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner. Nature 446, 820–823 (2007). https://doi.org/10.1038/nature05701
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature05701
This article is cited by
-
The Mediator complex as a master regulator of transcription by RNA polymerase II
Nature Reviews Molecular Cell Biology (2022)
-
Optical tweezers in single-molecule biophysics
Nature Reviews Methods Primers (2021)
-
Causes and consequences of RNA polymerase II stalling during transcript elongation
Nature Reviews Molecular Cell Biology (2021)
-
Single-molecule characterization of extrinsic transcription termination by Sen1 helicase
Nature Communications (2019)
-
Pause sequences facilitate entry into long-lived paused states by reducing RNA polymerase transcription rates
Nature Communications (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.