Abstract
Envelope glycoprotein (Env) spikes on AIDS retroviruses initiate infection of host cells and are therefore targets for vaccine development. Though crystal structures for partial Env subunits are known, the structure and distribution of native Env spikes on virions is obscure. We applied cryoelectron microscopy tomography to define ultrastructural details of spikes. Virions of wild-type human immunodeficiency virus 1 (HIV-1) and a mutant simian immunodeficiency virus (SIV) had ∼14 and ∼73 spikes per particle, respectively, with some clustering of HIV-1 spikes. Three-dimensional averaging showed that the surface glycoprotein (gp120) ‘head’ of each subunit of the trimeric SIV spike contains a primary mass, with two secondary lobes. The transmembrane glycoprotein ‘stalk’ of each trimer is composed of three independent legs that project obliquely from the trimer head, tripod-like. Reconciling available atomic structures with the three-dimensional whole spike density map yields insights into the orientation of Env spike structural elements and possible structural bases of their functions.
Similar content being viewed by others
Main
The Env spikes of HIV-1 are composed of surface envelope glycoprotein (gp120SU) and transmembrane envelope glycoprotein (gp41TM) subunits, which, in their native configuration, elicit partially effective humoral immune responses1,2 and represent obvious vaccine targets for viral prophylactics3,4,5,6. The Env spikes are thought to be trimeric7,8,9,10 and structure-based models have been proposed11,12,13. However, despite intensive efforts, the arrangement and orientation of the loop-deleted gp120 core atomic structures within a native Env spike and their association with the gp41 subunits have remained largely speculative12,13. We recently obtained electron tomograms of the Env spikes on intact, chemically fixed SIV and HIV-1, preserved in negative stain, which clearly reveal trimeric spikes on the viral surface14. Because of the staining, fixation and drying artefacts that can be associated with this method, some potentially important features of the virions and their associated trimers could not be deduced. We have now applied cryo-electron microscopy (cryoEM) tomography15 to investigate the surface features of unfixed, unstained frozen hydrated HIV-1 and SIV virions to reveal the distribution pattern of Env spikes on individual virions and to generate a three-dimensional (3D) model of the SIV Env spike. We have also attempted to reconcile this model of authentic virion trimers with available atomic structures of their constituent subunits.
Spike numbers and distribution
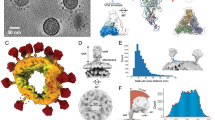
The cryoEM tomograms show that virions in our SIV and HIV-1 samples were intact and abundant with diameters of 109 ± 14 and 110 ± 8 nm, respectively (Fig. 1 and, for SIV, Supplementary Movie 1). Importantly, Env spikes are readily seen on virion surfaces (arrowheads). The SIV mac239 variant virus studied here has a mutation resulting in a transmembrane glycoprotein with a truncated cytoplasmic tail resulting in the elimination of endocytosis signals and a high rate of cell surface expression and viral incorporation16,17. CryoEM of the SIV virions (Fig. 1a, Fig. 2a and Supplementary Movie 1) showed a surface fairly well saturated with 73 ± 25 (n = 52) rather uniformly distributed Env spikes, in line with our previous data14. A nearest-neighbour analysis of interspike centre-to-centre spacing showed a distribution with a peak centring on ∼15 nm (Supplementary Fig. S1a). No evidence of discrete periodic peak distances was observed (Supplementary Fig. S1d) as might have been expected if spike placement reflected an underlying geometry of matrix/capsid structure.
Shown are computationally derived transverse sections through fields of SIV (a, left) and HIV-1 (b, left) virions and sequential Z-stack tomogram layers of selected virions (arrows on the left): a right, SIV Z-stack; b right, HIV-1 Z-stack. Examples of putative Env spikes on selected virions are indicated by arrowheads on the left. Scale bars, 100 nm.
In contrast to SIV, yet consistent with our earlier results14, cryoEM analysis of wild-type HIV-1 virions (n = 40), which have a native full-length gp41TM cytoplasmic tail, showed only 14 ± 7 Env spikes per particle (range 4 to 35) (see examples in Fig. 2b–d). As with SIV, no clear evidence of periodic spacing was observed (Supplementary Fig. S1e and f, respectively). The most frequent nearest-neighbour interspike spacing was ∼15 nm (Supplementary Fig. S1b) which was closer than was predicted (∼23 nm) from a virion model with randomly distributed spikes (see Supplementary Fig. S1c). A cluster analysis (Supplementary Fig. S2) also showed a higher proportion of closely spaced spikes than was predicted from chance alone (see examples in Fig. 2b and c). For example, 8.2% and 16.5% of the observed spikes were in clusters of at least seven members linked by distances of ≤22.5 nm and ≤30 nm, respectively. In contrast, 0.6% and 4.8% of the spikes were similarly linked in the random distribution model (Supplementary Fig. S2).
Greater numbers of spikes per virion are correlated with increased infectivity18. Fusion efficiency might also be dependent upon local spike density (clustering), especially if concerted receptor engagement of multiple Env spikes is required for binding and fusion10,19,20. Spike clustering may also facilitate antibody (Ab)-mediated neutralization by fostering bivalent cross-linking21,22,23. A paratope-to-paratope reach of 10–20 nm for typical Abs is well within the range of at least some epitopes on spikes 15 nm apart.
The clustering data are consistent with the hypothesis that Env spikes are not free to diffuse in the plane of the membrane. However, the lack of both an apparent ordered array of spikes and of evidence of periodicity in the inter-spike distances conflicts with the notion that the tails are inserted into a geometrically arrayed set of matrix pores24,25. Clustering need not reflect matrix association but could suggest a direct inter-spike interaction mediated by the cytoplasmic tails themselves.
The Env spike tomogram average and a spike model
We focused our initial cryoEM tomography efforts on the SIV virions with truncated transmembrane glycoproteins, as the significantly higher numbers of spikes per virion facilitated the analysis14. Though it seems unlikely, we cannot rule out the possibility that the lack of a full-length cytoplasmic tail has, in some way, influenced our cryoEM Env spike structure or that there could be significant structural differences between SIV and HIV-1 spikes.
To generate a model of the Env spike, 6,175 individual SIV spike volumes from 122 virions were aligned and averaged. On the basis of our earlier studies14 and the inherent three-fold symmetry in class averages of our cryoEM data obtained by correspondence analysis (Supplementary Methods and Fig. S3), the spike volume average was three-fold symmetrized to generate the final averaged trimer density map (Supplementary Fig. S4). Thus, each monomeric subunit within the final density map represents the data from 18,525 (that is, 6,175 × 3) individual monomeric subunits with a measured resolution of ∼3.2 nm. Eight unsymmetrized class averages (Supplementary Fig. S3) were not remarkably different from the three-fold symmetrized global average (Supplementary Fig. S4), suggesting a relatively homogeneous spike population at the resolution achieved.
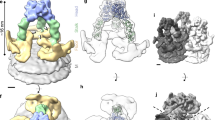
As shown in Fig. 3a–c and Supplementary Movie 2, each Env spike subunit contains three lobes, designated ‘main’, ‘lateral’ and ‘proximal’. Projecting from the three main lobes of the spike are ridges (‘arches’ in Fig. 3d) projecting medio-distally which join to form an apical ‘peak’ on the three-fold axis. The apical peak obscures an axial region of lower density; probably representing a hollow cavity just below it, a feature more apparent in a surface generated using a higher density threshold (Fig. 3d). Emanating from each proximal lobe is a short ‘leg’ that angles outward, whereupon it meets the viral ‘membrane’ and continues laterally just above the plane of the membrane to form a ‘foot’. The three splayed legs of the spike create a distinct cavity at the three-fold axis just above the membrane (Fig. 3a and e).
Env spike (blue) and membrane (grey) in side (a), intermediate (b), and top (c) views and an intermediate view at a higher density threshold (d). Panel e represents a membrane-parallel slice encompassing the legs and feet (MPER) as viewed from below. The centre-to-centre distance between possible MSD attachment sites (red lines) and the medial axis to MSD sites (green lines) are indicated. The spike and membrane surfaces (in a–d) were rendered at different thresholds, the latter adjusted to conform to the known thickness of the lipid bilayer (∼4 nm).
The height (13.7 nm) of the spike, the diameter of the head (10.5 nm), and the vertical height of the stalk (1.9 nm) are all in line with our previous measurements and most of those in the literature3,14. The diameter of each individual leg, a feature not previously described, ranges from about 1.9 to 3.7 nm. Because this region of the spike is believed to be rather flexible8, the features of the feet and legs in the model could be influenced by local density distortions resulting from averaging of these features when in different orientations.
The Moloney murine leukaemia virus (MMLV), the only other retroviral Env spike similarly analysed to date26, also shows three-fold symmetry. The MMLV Env spike appears to be squatter (∼10 × 10 nm) than the SIV spike—with multiple, more pronounced, distal and lateral projections26. Interestingly, the Env spikes on both viruses display an open tripod-like leg configuration. Thus, as discussed below, it appears that the membrane proximal regions of the three transmembrane glycoproteins of each trimer are not intimately associated in parallel in a combined stalk orthogonal to the viral envelope as is generally portrayed5,27.
The gp120 triad
There are now two 3D models of the gp120 trimer based on the crystal structures of variable loop-deleted AIDS virus gp120 core fragments: SIV in the pre-liganded presumptive native state conformation13, and HIV-1, liganded with CD4 and an anti-gp120 Fab fragment11,12. SIV and HIV-1 Env are considered similar enough in sequence to allow valid structural comparisons13. However, the trimer models vary considerably from one another, based largely on structural differences associated with large conformational changes induced by receptor binding. Consequently, only the unliganded SIV model may be appropriately compared to our unliganded SIV cryoEM density map.
An initial attempt to fit the unliganded SIV gp120 core trimer model (missing structures including the V1/V2 and V3 loops) of Chen et al. (figure 6 in ref. 13) into our averaged cryoEM Env spike density resulted in a considerable proportion of the atomic structure protruding from the periphery (Supplementary Fig. S5a). Efforts to move the protomers medially improved the fit somewhat but appeared sterically to restrict access to the disto-medially oriented CD4 binding sites (Supplementary Fig. S5b).
In a second fitting exercise (Fig. 4 and Supplementary Movie 3) we started afresh, attempting to optimize the positioning of the gp120 core structures (Fig. 4a, b, d–f) within the main lobes of the density map while maintaining orientation of the truncated amino and carboxy termini and the unstructured segment of the C2 loop downward and towards the central three-fold axis, where contact with gp41 presumably occurs13. This positioning minimized the loops and side chains protruding from our cryoEM envelope (Fig. 4a, d and Supplementary Fig. S5c) and positioned the CD4 binding sites on the periphery of the spike. The optimum fit aligned the truncated stems of the V1/V2 and V3 loops towards the otherwise unoccupied proximal and lateral lobes, respectively (Fig. 4a–e). The apical peak is shown unoccupied by the atomic model, consistent with its low density (Fig. 4f). The arch in Fig. 3d may represent an alternative position of the flexible V3 loop (that is, projecting towards the target cell surface) on a minority of the Env subunits, a conformation recently defined at the atomic level for the V3 loop in the CD4-bound fusion-active HIV-1 protomer28.
Atomic model: Protein Data Bank ID: 2BF1 (ref. 13). Side (a, b) and top (d, e), views, with (a, d) and without (b, e) enshrouding cryoEM surface. Indicated structures: gp120 subunits (blue, magenta, mauve), CD4 binding sites (red), V3 loop stems (yellow), V1/V2 loop stems (orange), N and C termini (green), C2 loop stems (white), proposed V3 and V1/V2 loop sites (mesh-enclosed volumes over stems or (in c) coloured surfaces). f, Spike as in a, but tilted and at higher density threshold, showing low-density region under peak.
The transmembrane glycoprotein
The fusion-active conformation of the transmembrane glycoprotein trimer is believed to be a rod-like extended coiled coil dominated by two α-helical regions, the N-terminal heptad repeat 1 (HR1) and C-terminal heptad repeat 2 (HR2) within each subunit. In contrast, the post-fusion conformation is defined by the 6-helix bundle wherein the HR2 regions have folded back on the HR1 regions, presumably destabilizing and drawing the viral and cell membranes together in the process7,8,9,29. The prefusion structure of the transmembrane glycoprotein is considerably less well defined30. Ab binding studies demonstrate that the region encompassed by HR1 and HR2 is essentially obscured in the native Env spike and so is not likely to contribute directly to the surface features of the Env spike30. This then leaves the membrane proximal external region (MPER: the segment between HR2 and the membrane-spanning domain, MSD) as a potential target for neutralizing Ab and the probable contributor to the bulk of the Env stalk observed by cryoEM.
The MPER is the most highly conserved region in the transmembrane glycoprotein27 and has been proposed to serve as (1) a flexible extender8, (2) a contributor to trimer formation and stability31,32, and (3) an agent for membrane destabilization that fosters membrane fusion27. The amphipathic nature of the C-terminal portions of the MPER and the tendency for peptides of this region to self-associate have been interpreted as indicating that this region makes an important contribution to Env spike trimerization31. Our data contradict this notion. The lower density that we observe between the legs of the stem region, which appears as a pronounced gap in the surface-rendered model (Fig. 3a, e) indicates that this region of the transmembrane glycoprotein subunits is not self-associated in native trimers on prefusion virions but rather is engaged in an extensive interaction with the plasma membrane. In support of this interpretation are the observations that the anti-MPER monoclonal Abs (MAbs) 2F5 and 4E10 bind better to their respective epitopes when the epitopes are associated with lipid33,34. Moreover, the 2F5 and 4E10 epitopes and, remarkably, the tips of the 2F5 and 4E10 MAbs themselves, display pronounced hydrophobic patches34,35.
We manually mapped the 2F5 and 4E10 peptides into our SIV spike model under the assumption that this highly conserved region would be similarly configured in HIV-1 and SIV. In support of this assumption, both epitopes, when grafted into SIV, have been shown to be immunologically indistinguishable from those in HIV-1 (ref. 36). The placement of the 2F5 and 4E10 peptides in our map was facilitated by positioning the relevant hydrophobic residues of the peptides and their respective MAbs in the plane of the membrane. We found that the splayed legs of the tomogram model readily accommodated the peptides and are consistent with the cognate MAbs pointing towards the membrane (rather than towards a compact stalk) (Fig. 5 and Supplementary Movie 4).
Surface-rendered model of the SIV Env spike showing proposed placement of 2F5 (blue; PDB ID: 1TJI) and 4E10 (red; PDB ID: 1TZG) HIV-1 epitope peptides in the leg and foot densities (a) based on alignment of hydrophobic residues in the peptides and Abs parallel to the membrane. The proposed positions of the corresponding antigen-binding fragment (Fab) 2F5 (turquoise, b) and Fab 4E10 (pink, c), are shown. Red cylinders indicate proposed positions of the helical MSDs relative to the spike feet.
The hydrophobicity of the MPER is a highly conserved feature of SIV, MMLV, HIV, and other retroviruses. Both SIV and MMLV, the two retroviruses for which cryoEM data are available, display splayed legs wherein the proximal regions seem to be associated with the membrane. Within HIV-1, the 4E10 epitope may be universally expressed37, implying a strong conservation of function for this region. Together, these observations suggest a critical role for an association between the MPER and the outer leaflet of the viral membrane in viral function.
The MSD and matrix association
The MSD is generally believed to be an α-helical region composed primarily of neutral and hydrophobic amino acids38 which serves to stabilize the trimer by self-association. However, steric considerations, as outlined above, suggest that the MSDs are most probably displaced a considerable distance (∼4.3 nm) from the three-fold axis (Figs 3a, e and 5a) with a likely centre-to-centre distance of ∼8 nm. Consequently, any contribution to trimer stabilization in the prefusion configuration is probably due to simple membrane anchorage. It has been suggested that triggering of a single retrovirus spike is sufficient to initiate membrane fusion39, in contrast to the engagement of multiple spikes as, for example, is believed necessary for influenza virus fusion40,41. If so, then perhaps the three splayed legs with their unassociated MSDs, in conjunction with membrane-associated elements of the MPERs, provide the added degree of lipid anchorage required by some aspect of the fusion mechanism. One possible mechanism would be the fostering of more efficient and extensive recruitment and disruption of the outer leaflet of the viral membrane by the three widely spaced MPER/MSD structures of a single spike during the large conformational change in the Env spike that occurs upon triggering. In addition to mechanistic considerations, the open leg configuration could have important implications for the designs of future soluble Env trimer vaccine candidates, particularly those stabilized by engineered trimerization motifs.
Though we do not see evidence of it in mature virions, it is believed that the viral matrix proteins both interact with the cytoplasmic tails of Env and are organized into a regular geometric array24,42. Nevertheless, the completeness of the array has been questioned. Models of matrix protein arrays, in which the resulting holes are spaced at about 7 nm intervals, have been proposed24,43. Such patterns could accommodate 8 nm centre-to-centre spacing for MSDs if the cytoplasmic tails are angled slightly towards the three-fold axis.
Finally, it must be noted that only a minority of HIV-1 or SIV virions in typical preparations possess culturable infectivity, and it has been proposed that there may be non-functional forms of Env present on the virion surface44. It is thus possible that the trimers of infectious particles may differ structurally from the averaged structures described here, although there are several factors that contribute to the low particle infectivity ratio of HIV-1 and SIV, many of which are unrelated to the viral Env. In any case, the current results provide a new level of structural information about native trimers on virions, and guidance for further studies.
Discussion
The present studies begin to bridge the gap between atomic level structures derived from crystallographic studies of partial Env spike subunits and the resolution of conventional transmission electron microscopy, revealing 3D ultrastructural details of individual native trimers. Additional studies of virions with variable loop deletions and of virions complexed with Env-targeted ligands of structurally defined specificity should provide additional insights relevant to further understanding of virus binding and fusion and Ab neutralization.
Methods
More detailed Methods are available as Supplementary Information.
Viruses
Highly purified virus samples45 treated with Aldrithiol-2 (ref. 46) were provided by the AIDS Vaccine Program (SAIC Frederick, NCI, Maryland, USA).
CryoEM preparation
Virus aliquots were washed and placed on EM grids for 1 min. Excess virus and buffer were removed by blotting, and the sample rapidly vitrified. The grids were either examined immediately or stored in liquid nitrogen for later use.
Cryo-electron tomography
The EM grids were examined under low-dose conditions. Single axis tilt series of 70–80 images were recorded at 43,200 × magnification and an underfocus of 4–6 µm.
Image analysis
Tomograms were computed by weighted back-projection using the PROJALIGN program package47 and imported into the software package AMIRA for visualization and surface rendering.
SIV spike volumes, manually selected on the basis of appearances, size and location, were subjected to 3D alignment and averaging algorithms48. The morphologically most divergent 2% were excluded. The remaining spikes were aligned against a three-fold symmetrized reference and averaged. The average was then three-fold symmetrized to generate the final averaged trimer density map (Supplementary Fig. S4). The Fourier transform extended to 2.5 nm by Fourier Shell Correlation. The minimum distance measured between discrete internal density peaks was 3.2 nm (Supplementary Fig. S6).
Atomic model fitting
The indicated X-ray structures were manually fitted into the 3D averaged Env spike CryoEM density map using the Chimera Software package (http://www.cgl.ucsf.edu/chimera/).
Env spike distribution analyses
Neighbouring spikes within 22.5 nm or 30 nm were considered members of a cluster that included all other spikes similarly linked. The observed spike distribution data were compared with mathematically generated random distributions as described in Supplementary Fig. S1.
References
Wei, X. et al. Antibody neutralization and escape by HIV-1. Nature 422, 307–312 (2003)
Richman, D. D., Wrin, T., Little, S. J. & Petropoulos, C. J. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl Acad. Sci. USA 100, 4144–4149 (2003)
Poignard, P., Saphire, E. O., Parren, P. & Burton, D. Gp120: biologic aspects of structural features. Annu. Rev. Immunol. 19, 253–274 (2001)
Parren, P. W. & Burton, D. R. The antiviral activity of antibodies in vitro and in vivo. Adv. Immunol. 77, 195–262 (2001)
Burton, D. R. et al. HIV vaccine design and the neutralizing antibody problem. Nature Immunol. 5, 233–236 (2004)
Mascola, J. R. et al. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J. Virol. 73, 4009–4018 (1999)
Chan, D. C., Fass, D., Berger, J. M. & Kim, P. S. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89, 263–273 (1997)
Weissenhorn, W., Dessen, A., Harrison, S. C., Skehel, J. J. & Wiley, D. C. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387, 426–430 (1997)
Caffrey, M. et al. Three-dimensional solution structure of the 44 kDa ectodomain of SIV gp41. EMBO J. 17, 4572–4584 (1998)
Doms, R. W. Beyond receptor expression: the influence of receptor conformation, density, and affinity in HIV-1 infection. Virology 276, 229–237 (2000)
Kwong, P. D. et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648–659 (1998)
Kwong, P. D., Wyatt, R., Sattentau, Q. J., Sodroski, J. & Hendrickson, W. A. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J. Virol. 74, 1961–1972 (2000)
Chen, B. et al. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433, 834–841 (2005)
Zhu, P. et al. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc. Natl Acad. Sci. USA 100, 15812–15817 (2003)
Lucic, V., Forster, F. & Baumeister, W. Structural studies by electron tomography: from cells to molecules. Annu. Rev. Biochem. 74, 833–865 (2005)
LaBranche, C. C. et al. A single acid change in the cytoplasmic domain of the simian immunodeficiency virus transmembrane molecule increases envelope glycoprotein expression on infected cells. J. Virol. 69, 5217–5227 (1995)
Kodama, T. et al. Significance of premature stop codons in env of simian immunodeficiency virus. J. Virol. 63, 4709–4714 (1989)
Yuste, E., Reeves, J. D., Doms, R. W. & Desrosiers, R. C. Modulation of Env content in virions of simian immunodeficiency virus: correlation with cell surface expression and virion infectivity. J. Virol. 78, 6775–6785 (2004)
Doms, R. W. & Moore, J. P. HIV-1 membrane fusion: targets of opportunity. J. Cell Biol. 151, F9–F13 (2000)
Kuhmann, S. E., Platt, E. J., Kozak, S. L. & Kabat, D. Cooperation of multiple CCR5 coreceptors is required for infections by human immunodeficiency virus type 1. J. Virol. 74, 7005–7015 (2000)
Cavacini, L. A., Emes, C. L., Power, J., Duval, M. & Posner, M. R. Effect of antibody valency on interaction with cell-surface expressed HIV-1 and viral neutralization. J. Immunol. 152, 2538–2545 (1994)
Klasse, P. J. & Sattentau, Q. J. Mechanisms of virus neutralization by antibody. Curr. Top. Microbiol. Immunol. 260, 87–108 (2001)
Yuste, E., Johnson, W., Pavlakis, G. N. & Desrosiers, R. C. Virion envelope content, infectivity, and neutralization sensitivity of simian immunodeficiency virus. J. Virol. 79, 12455–12463 (2005)
Forster, M. J., Mulloy, B. & Nermut, M. V. Molecular modelling study of HIV p17gag (MA) protein shell utilising data from electron microscopy and X-ray crystallography. J. Mol. Biol. 298, 841–857 (2000)
Hill, C. P., Worthylake, D., Bancroft, D. P. & Christensen, A. M. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: Implications for membrane association and assembly. Proc. Natl Acad. Sci. USA 93, 3099–3104 (1996)
Förster, F., Medalia, O., Zauberman, N., Baumeister, W. & Fass, D. Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography. Proc. Natl Acad. Sci. USA 102, 4729–4734 (2005)
Zwick, M. B. et al. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75, 10892–10905 (2001)
Huang, C. et al. Structure of a V3-containing HIV-1 gp120 core. Science 310, 1025–1028 (2005)
Tan, K., Liu, J.-H., Wang, J.-H., Shen, S. & Lu, M. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl Acad. Sci. USA 94, 12303–12308 (1997)
Qiao, Z. et al. Design, expression, and immunogenicity of a soluble HIV trimeric envelope fragment adopting a prefusion gp41 configuration. J. Biol. Chem. 280, 23138–23146 (2005)
Salzwedel, K., West, J. T. & Hunter, E. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J. Virol. 73, 2469–2480 (1999)
Saez-Cirion, A. et al. Structural and functional roles of HIV-1 gp41 pretransmembrane sequence segmentation. Biophys. J. 85, 3769–3780 (2003)
Grundner, C., Mirzabekov, T., Sodroski, J. & Wyatt, R. Solid-phase proteoliposomes containing human immunodeficiency virus envelope glycoproteins. J. Virol. 76, 3511–3521 (2002)
Ofek, G. et al. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J. Virol. 78, 10724–10737 (2004)
Cardoso, R. M. et al. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity 22, 163–173 (2005)
Yuste, E. et al. Simian immunodeficiency virus engrafted with human immunodeficiency virus type 1 (HIV-1)-specific epitopes: replication, neutralization, and survey of HIV-1-positive plasma. J. Virol. 80, 3030–3041 (2006)
Binley, J. M. et al. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78, 13232–13252 (2004)
West, J. T., Johnston, P. B., Dubay, S. R. & Hunter, E. Mutations within the putative membrane-spanning domain of the simian immunodeficiency virus transmembrane glycoprotein define the minimal requirements for fusion, incorporation, and infectivity. J. Virol. 75, 9601–9612 (2001)
Yang, X., Kurteva, S., Ren, X., Lee, S. & Sodroski, J. Stoichiometry of envelope glycoprotein trimers in the entry of human immunodeficiency virus type 1. J. Virol. 79, 12132–12147 (2005)
Blumenthal, R., Sarkar, D. P., Durell, S., Howard, D. E. & Morris, S. J. Dilation of the influenza hemagglutinin fusion pore revealed by the kinetics of individual cell-cell fusion events. J. Cell Biol. 135, 63–71 (1996)
Danieli, T., Pelletier, S. L., Henis, Y. I. & White, J. M. Membrane fusion mediated by the influenza virus hemagglutinin requires the concerted action of at least three hemagglutinin trimers. J. Cell Biol. 133, 559–569 (1996)
Freed, E. O. & Martin, M. A. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J. Virol. 70, 341–351 (1996)
Rao, Z. et al. Crystal structure of SIV matrix antigen and implications for virus assembly. Nature 378, 743–747 (1995)
Moore, P. L. et al. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J. Virol. 80, 2515–2528 (2006)
Chertova, E. et al. Sites, mechanism of action and lack of reversibility of primate lentivirus inactivation by preferential covalent modification of virion internal proteins. Curr. Mol. Med. 3, 265–272 (2003)
Rossio, J. L. et al. Inactivation of human immunodeficiency virus type 1 infectivity with preservation of conformational and functional integrity of virion surface proteins. J. Virol. 72, 7992–8001 (1998)
Winkler, H. & Taylor, K. A. Accurate marker-free alignment with simultaneous geometry determination and reconstruction of tilt series in electron tomography. Ultramicroscopy 106, 240–254 (2006)
Winkler, H. & Taylor, K. A. Multivariate statistical analysis of three-dimensional cross-bridge motifs in insect flight muscle. Ultramicroscopy 77, 141–152 (1999)
Acknowledgements
We thank H. Winkler for assistance with volume alignment and classification and 3D volume pickup and P. D. Kwong for his comments on the manuscript. The work was supported in part by a NIH and NCI contract (J.B., E.C., J.D.L.) and grants from the NIH NIAID (P.Z., H.G., K.H.R.) and the NIH NIGMS (J.L., K.A.T.). This project has been funded in whole or in part with federal funds from the National Cancer Institute, NIH. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.Author Contributions P.Z. performed the bulk of the cryoEM, image analysis, model fitting, and graphic illustration. J.L. assisted with the image acquisition, analysis and interpretation. J.B. cultured and purified the viruses. E.C. biochemically analysed the virus preparations for purity. J.D.L. contributed to the design of the project and contributed extensively to the manuscript writing. H.G. assisted with fitting of the 2F5 and 4E10 peptides and MAbs into the cryoEM density map. G.A.O. contributed to the analysis of the data, particularly as it relates to the transmembrane region of the envelope spike. K.A.T. provided technical expertise on various aspects of the cryoEM and tomography and contributed to technical portions of the manuscript. K.H.R. provided overall management of the project as well as data interpretation, manuscript writing and figure design.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Notes
This file contains Supplementary Methods, Supplementary Figure Legends, and Supplementary Movie Legends and additional references. (DOC 46 kb)
Supplementary Figure 1
Distribution of inter-spike centre-to-centre distances for mutant SIV and wtHIV-1. (JPG 78 kb)
Supplementary Figure 2
Distribution of Env spike cluster sizes on 40 HIV-1 virions compared with the distribution of clusters on hypothetical model virions with randomly placed Env spikes calculated, as described for Supplementary Figure 1. (JPG 45 kb)
Supplementary Figure 3
Inherent three-fold symmetry in Env spike averaged images. (JPG 76 kb)
Supplementary Figure 4
Z-stack of 42 sequential density sections through the 3-fold axis of the final Env spike average. (JPG 77 kb)
Supplementary Figure 5
Comparison of gp120 fitted trimer models. (JPG 148 kb)
Supplementary Figure 6
Density distribution profile of the cryoEM trimer slice through the final Env spike average showing the minimum separation between distinguishable internal density peaks measuring 3.2 nm. (JPG 44 kb)
Supplementary Movie 1
CryoEM tomogram of SIV virions and corresponding isosurface model. (MOV 9749 kb)
Supplementary Movie 2
CryoEM spike Env model. (MOV 9115 kb)
Supplementary Movie 3
De novo fitting of gp120 atomic model into cryoEM isosurface model. (MOV 9282 kb)
Supplementary Movie 4
CryoEM isosurface model manually fitted with gp41 peptides and docked with MAbs. (MOV 8508 kb)
Rights and permissions
About this article
Cite this article
Zhu, P., Liu, J., Bess, J. et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441, 847–852 (2006). https://doi.org/10.1038/nature04817
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature04817
This article is cited by
-
Nanoparticles exhibiting virus-mimic surface topology for enhanced oral delivery
Nature Communications (2023)
-
Inadequate structural constraint on Fab approach rather than paratope elicitation limits HIV-1 MPER vaccine utility
Nature Communications (2023)
-
Directing HIV-1 for degradation by non-target cells, using bi-specific single-chain llama antibodies
Scientific Reports (2022)
-
Phagocytosis by an HIV antibody is associated with reduced viremia irrespective of enhanced complement lysis
Nature Communications (2022)
-
Neutralizing antibody responses to SARS-CoV-2 in symptomatic COVID-19 is persistent and critical for survival
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.