Abstract
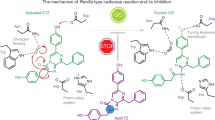
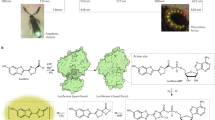
Fireflies communicate with each other by emitting yellow-green to yellow-orange brilliant light. The bioluminescence reaction, which uses luciferin, Mg-ATP and molecular oxygen to yield an electronically excited oxyluciferin species, is carried out by the enzyme luciferase. Visible light is emitted during relaxation of excited oxyluciferin to its ground state. The high quantum yield1 of the luciferin/luciferase reaction and the change in bioluminescence colour caused by subtle structural differences in luciferase have attracted much research interest. In fact, a single amino acid substitution in luciferase changes the emission colour from yellow-green to red2,3,4,5. Although the crystal structure of luciferase from the North American firefly (Photinus pyralis) has been described6,7, the detailed mechanism for the bioluminescence colour change is still unclear8,9,10,11. Here we report the crystal structures of wild-type and red mutant (S286N) luciferases from the Japanese Genji-botaru (Luciola cruciata) in complex with a high-energy intermediate analogue, 5′-O-[N-(dehydroluciferyl)-sulfamoyl]adenosine (DLSA). Comparing these structures to those of the wild-type luciferase complexed with AMP plus oxyluciferin (products) reveals a significant conformational change in the wild-type enzyme but not in the red mutant. This conformational change involves movement of the hydrophobic side chain of Ile 288 towards the benzothiazole ring of DLSA. Our results indicate that the degree of molecular rigidity of the excited state of oxyluciferin, which is controlled by a transient movement of Ile 288, determines the colour of bioluminescence during the emission reaction.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Seliger, H. H. & McElroy, W. D. Spectral emission and quantum yield of firefly bioluminescence. Arch. Biochem. Biophys. 88, 136–141 (1960)
Kajiyama, N. & Nakano, E. Isolation and characterization of mutants of firefly luciferase which produce different colors of light. Protein Eng. 4, 691–693 (1991)
Mamaev, S. V., Laikhter, A. L., Arslan, T. & Hecht, S. M. Firefly luciferase: Alteration of the color of emitted of light resulting from substitutions at position 286. J. Am. Chem. Soc. 118, 7243–7244 (1996)
Branchini, B. R., Southworth, T. L., Murtiashaw, M. H., Boije, H. & Fleet, S. E. A mutagenesis study of the putative luciferin binding site residues of firefly luciferase. Biochemistry 42, 10429–10436 (2003)
Branchini, B. R. et al. Site-directed mutagenesis of firefly luciferase active site amino acids: a proposed model for bioluminescence color. Biochemistry 38, 13223–13230 (1999)
Conti, E., Franks, N. P. & Brick, P. Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure 4, 287–298 (1996)
Franks, N. P., Jenkins, A., Conti, E., Lieb, W. R. & Brick, P. Structural basis for the inhibition of firefly luciferase by a general anesthetic. Biophys. J. 75, 2205–2211 (1998)
DeLuca, M. Hydrophobic nature of the active site of firefly luciferase. Biochemistry 8, 160–166 (1969)
White, E. H., Rapaport, E., Hopkins, T. A. & Seliger, H. H. Chemi- and bioluminescence of firefly luciferin. J. Am. Chem. Soc. 91, 2178–2180 (1969)
Branchini, B. R. et al. An alternative mechanism of bioluminescence color determination in firefly luciferase. Biochemistry 43, 7255–7262 (2004)
McCapra, F., Gilfoyle, D. J., Young, D. W., Church, N. J. & Spencer, P. in Bioluminescence and Chemiluminescence: Fundamental and Applied Aspects (eds Campbell, A. K., Krick, L. J. & Stanley, P. E.) 387–391 (John Wiley and Sons, Chichester, 1994)
Deluca, M. Firefly luciferase. Adv. Enzymol. 44, 37–68 (1976)
Delarue, M. Aminoacyl-tRNA synthetases. Curr. Opin. Struct. Biol. 5, 48–55 (1995)
Chang, K. H., Xiang, H. & Dunaway-Mariano, D. Acyl-adenylate motif of the acyl-adenylate/thioester-forming enzyme superfamily: a site-directed mutagenesis study with the Pseudomonas sp. strain CBS3 4-chlorobenzoate:coenzyme A ligase. Biochemistry 36, 15650–15659 (1997)
Kleinkauf, H. & Von Dohren, H. A nonribosomal system of peptide biosynthesis. Eur. J. Biochem. 236, 335–351 (1996)
Babbitt, P. C. et al. Ancestry of the 4-chlorobenzoate dehalogenase: analysis of amino acid sequence identities among families of acyl:adenyl ligases, enoyl-CoA hydratases/isomerases, and acyl-CoA thioesterases. Biochemistry 31, 5594–5604 (1992)
May, J. J., Kessler, N., Marahiel, M. A. & Stubbs, M. T. Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases. Proc. Natl Acad. Sci. USA 99, 12120–12125 (2002)
Ueda, H. et al. X-ray crystallographic conformational study of 5′-O-[N-(l-alanyl)-sulfamoyl]adenosine, a substrate analogue for alanyl-tRNA synthetase. Biochim. Biophys. Acta 1080, 126–134 (1991)
Branchini, B. R., Murtiashaw, M. H., Carmody, J. N., Mygatt, E. E. & Southworth, T. L. Synthesis of an N-acyl sulfamate analog of luciferyl-AMP: a stable and potent inhibitor of firefly luciferase. Bioorg. Med. Chem. Lett. 15, 3860–3864 (2005)
Conti, E., Stachelhaus, T., Marahiel, M. A. & Brick, P. Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. EMBO J. 16, 4174–4183 (1997)
Kumasaka, T., Yamamoto, M., Yamashita, E., Moriyama, H. & Ueki, T. Trichromatic concept optimizes MAD experiments in synchrotron X-ray crystallography. Structure 10, 1205–1210 (2002)
Adachi, S. et al. The RIKEN structural biology beamline II (BL44B2) at the SPring-8. Nucl. Instrum. Methods A 467–468, 711–714 (2001)
Pflugrath, J. W. The finer things in X-ray diffraction data collection. Acta Crystallogr. D 55, 1718–1725 (1999)
Leslie, A. G. W. in Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography No. 26 (CCP4, Warrington, 1992)
Navaza, J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A 50, 157–163 (1994)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Perrakis, A., Morris, R. & Lamzin, V. S. Automated protein model building combined with iterative structure refinement. Nature Struct. Biol. 6, 458–463 (1999)
Roussel, A. & Cambillau, C. Turbo-Frodo Manual (AFMB-CNRS, Marseille, 1996)
DeLano, W. L. The PyMOL Molecular Graphics System. (DeLano Scientific, San Carlos, California, 2002)
Acknowledgements
We thank N. Kajiyama and K.Hirokawa (Kikkoman Corp.) for providing cDNA of luciferase and measurement of bioluminescence spectra. We also thank M. Yamamoto and S. Adachi (RIKEN Harima Institute) for X-ray diffraction data collection on the RIKEN beamlines BL45XU and BL44B2 at SPring-8. This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (T.N. and H.K.)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Atomic coordinates for luciferase structures have been deposited in the Protein Data Bank under accession codes 2D1Q (WT·Mg-ATP), 2D1S (WT·DLSA), 2D1R (WT·AMP/oxyluciferin) and 2D1T (S286N·DLSA). The authors declare no competing financial interests.
Supplementary information
Supplementary Notes
This file contains Supplementary Discussion, Supplementary Methods, Supplementary Figure Legends, Supplementary Movie Legends and Supplementary Table 1. (DOC 111 kb)
Supplementary Figures
This file contains Supplementary Figures 1–3. (PDF 1320 kb)
Supplementary Movie 1
A movie of the luminescence reaction of wild-type luciferase. (MOV 427 kb)
Supplementary Movie 2
A movie of the luminescence reaction of S286N mutant luciferase. (MOV 432 kb)
Rights and permissions
About this article
Cite this article
Nakatsu, T., Ichiyama, S., Hiratake, J. et al. Structural basis for the spectral difference in luciferase bioluminescence. Nature 440, 372–376 (2006). https://doi.org/10.1038/nature04542
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature04542
This article is cited by
-
Role of Histidine 310 in Amydetes vivianii firefly luciferase pH and metal sensitivities and improvement of its color tuning properties
Photochemical & Photobiological Sciences (2024)
-
The orange light emitting luciferase from the rare Euryopa clarindae adult railroadworm (Coleoptera:Phengodidae): structural/functional and evolutionary relationship with green and red emitting luciferases
Photochemical & Photobiological Sciences (2024)
-
Practical Guidance for Developing Small-Molecule Optical Probes for In Vivo Imaging
Molecular Imaging and Biology (2023)
-
Rapid Detection of Hepatitis A Virus in Foods Using a Bioluminescent Assay in Real-Time (BART) and Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP) Technology
Food and Environmental Virology (2023)
-
Effect of pH on the secondary structure and thermostability of beetle luciferases: structural origin of pH-insensitivity
Photochemical & Photobiological Sciences (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.