Abstract
Male breast cancer is a rare disease that is still poorly understood. It is mainly classified by immunohistochemistry as a luminal disease. In this study, we assess for the first time the correlation between molecular subtypes based on a validated six-marker immunohistochemical panel and PAM50 signature in male breast cancer, and the subsequent clinical outcome of these different subtypes. We collected 67 surgical specimens of invasive male breast cancer from four different Spanish pathology laboratories. Immunohistochemical staining for the six-marker panel was performed on tissue microarrays. PAM50 subtypes were determined in a research-use-only nCounter Analysis System. We explored the association of immunohistochemical and PAM50 subtypes. Overall survival and disease-free survival were analyzed in the different subtypes of each classification. The distribution of tumor molecular subtypes according PAM50 was: 60% luminal B, 30% luminal A and 10% human epidermal growth factor receptor 2 (Her2) enriched. Only one Her2-enriched tumor was also positive by immunohistochemistry and was treated with trastuzumab. None of the tumors were basal-like. Using immunohistochemical surrogates, 51% of the tumors were luminal B, 44% luminal A, 4% triple-negative and 1% Her2-positive. The clinicopathological characteristics did not differ significantly between immunohistochemical and PAM50 subtypes. We found a significant worse overall survival in Her2-enriched compared with luminal tumors. Male breast cancer seems to be mainly a genomic luminal disease with a predominance of the luminal B subtype. In addition, we found a proportion of patients with Her2-negative by immunohistochemistry but Her2-enriched profile by PAM50 tumors with a worse outcome compared with luminal subtypes that may benefit from anti-Her2 therapies.
Similar content being viewed by others
Main
Male breast cancer is a rare disease, accounting for <1% all breast cancer diagnoses.1 In general, the prognosis for males and females with breast cancer is similar. Overall survival rates are lower for men, but this is due to an older age at diagnosis, resulting in higher levels of comorbidity and more advanced stages at presentation.2 As a result of the low incidence of male breast cancer and the lack of data from prospective randomized trials, its medical treatment is based largely on evidence from studies with small numbers of patients or on studies of female breast cancer.
Breast cancer is a heterogeneous disease. In recent years, discoveries facilitated by molecular biology in female breast cancer have allowed us to move from a purely anatomical and pathological classification to a new classification based on molecular criteria.3 Immunohistochemical markers have been used as surrogates for DNA microarray in determining breast cancer subtypes. In this way, it has been demonstrated that the outcome value of the basal-like subtype is significantly better when determined by five immunohistochemical markers (estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (Her2), epidermal growth factor receptor (EGFR) and cytokeratin 5/6 (CK5/6)) compared with a triple-negative definition (ER, PR and Her2).4, 5, 6 However, although an immunohistochemical staining proxy can be used to stratify and classify breast cancers in a clinical setting, it cannot fully reproduce gene expression classification. This situation was illustrated by Rivenbark et al7 in 381 breast cancers for which both immunohistochemical and molecular gene expression were determined. The ER-positive/PR-positive/Her2-negative subset contained 90% of the luminal A tumors by gene expression, but these represented only 49% of the tumors classified as ER-positive/PR-positive/Her2-negative.
Perou et al3 studied the biological diversity of breast cancer through the identification of intrinsic breast cancer subtypes with gene expression patterns using DNA microarrays.8 Five subtypes were recognized, two of which were derived from ER-positive tumors (luminal A and B) and three of which were derived from ER-negative tumors (normal breast like, basal and Her2-enriched).8, 9 These molecular differences appeared to be associated with both the clinical outcome and the response to chemotherapy. Several gene signatures are currently being used to predict outcomes in breast cancer. PAM50 gene signature is a second-generation multigene expression assay to quantify mRNA expression of 50 genes, including probes for ER, PR and Her2. PAM50 signature can classify tumor samples into the four intrinsic subtypes and provides an accurate estimate of the risk of distant recurrence in hormone receptor-positive female breast cancer when analyzed with the Prosigna® algorithm.10, 11 Despite growing evidence of biological differences between male and female breast cancer, currently there are no data on male breast cancer molecular subtyping based on gene signatures, including the PAM50 classification of female breast cancer. The aim of this study is to report, for the first time, the correlation between molecular subtypes based on a validated six-marker immunohistochemical panel and PAM50 signature in male breast cancer, and the subsequent clinical outcome of the different subtypes.
Materials and methods
Patients and Samples
Between 1996 and 2015, surgical specimens of invasive male breast cancer were collected from four different Spanish pathology laboratories (Hospitales Universitarios Regional y Virgen de la Victoria, Málaga; Complejo Hospitalario de Jaén, Jaén; Hospital de la Serranía, Ronda; and Hospital Costa del Sol, Marbella). Medical records libraries were reviewed in order to retrieve clinical information and follow-up. Samples were managed and provided by the Málaga Hospital-IBIMA Biobank that belongs to the Andalusian Public Health System Biobank, belonging to the National Biobank Platform (project PT13/0010/0006 and PT13/0010/000013). All patients participating in the study gave their informed consent and protocols were approved by institutional ethical committees (Comité Coordinador de Ética de la Investigación Biomédica de Andalucía).
Immunohistochemical Subtyping
Formalin-fixed, paraffin-embedded tissue blocks were retrieved from the above-mentioned hospitals. Hematoxylin and eosin-stained slides were used to select representative tissue blocks with the greatest area of viable invasive breast carcinoma and identify tumor areas. From these areas, three cores sized 0.6-mm were obtained to build tissue microarrays with a manual tissue arrayer (Beecher Instruments, Sun Prairie, WI, USA). Immunohistochemical staining was performed on 5 μm sections from tissue microarray blocks and carried out in an automatic immunostainer (Autostainer Plus, Dako, Glostrup, Denmark) using the EnVision FLEX System (Dako). Diaminobenzidine was used as chromogen, counterstained with hematoxylin. As a negative control, the primary antibody was replaced by a nonimmune serum, and positive controls were specific to each antibody. The following antibodies were used: ER (rabbit monoclonal antibody, prediluted, clone SP1; Master Diagnostica), PR (rabbit monoclonal antibody, prediluted, clone Y85; Master Diagnostica), CK5/6 (mouse monoclonal, prediluted, clone D5/16B4; Master Diagnostica), EGFR (rabbit monoclonal antibody, prediluted, clone EP38Y; Master Diagnostica), Her2 (Kit Herceptest Dako), androgen receptor AR (mouse monoclonal antibody, prediluted, clone AR441; Dako) and Ki-67 (rabbit monoclonal antibody, prediluted, clone SP6; Master Diagnostica).
The immunohistochemical scoring was conducted by two experienced pathologists (LV and LP-V) independently and blinded to other features. Positivity for ER, PR and AR was defined as any nuclear staining (>1%).12 EGFR and CK5/6 were considered positive if any membranous or cytoplasmic staining was observed in the invasive carcinoma. To determine Ki-67 index, the percentage of nuclear-positive neoplastic cells was quantified. Immunohistochemical assessment of Her2 was performed according to the recommendations of the guidelines published in 2013.13 In cases of Her2-positive +2, we investigated the gene amplification by chromogenic in situ hybridization (CISH) with Her2 CISH Kit pharm Dx (Dako).
The immunohistochemical expression profile of each tumor was used to classify them in the following molecular subtypes: luminal A (ER-positive and/or PR-positive, Her2-negative and Ki-67 <14%), luminal B (ER-positive and/or PR-positive, Her2-negative and Ki-67 ≥14%), Her2-positive (Her2-positive, independently of ER and PR status), basal-like (ER, PR and Her2-negative, and EGFR-positive and/or CK5/6-positive) and non-basal triple-negative (ER, PR, Her2, EGFR and CK5/6-negative).5, 6
PAM50 Subtyping
From enriched tumor areas (>10% tumor cellularity), 3–6 sections of 10 μm were obtained for RNA extraction and purification. It was conducted using the High Pure FFPET RNA Isolation Kit (Roche Life Science) according NanoString Technologies guidelines. Optical density of total RNA was measured at 260 and 280 nm to determine yield and purity using a low-volume spectrophotometer (Nanodrop, Thermo Scientific). RNA samples passed quality control if the measured concentration was ≥12.5 ng/μl and the A260/280 ratio was 1.7–2.5. Gene expression profiling was performed on a research-use-only (RUO) nCounter Analysis System using the RUO PAM50 assay according to the manufacturer’s guidelines. Data were analyzed with the Prosigna algorithm by Nanostring Technologies team.
Statistical Methods
Data analysis was performed using R statistical language, base package version 3.3.0 and the CRAN-distributed packages Survival (version 2.38) and gmodels (version 2.16.2), with a general descriptive analysis of the variables included in the study. Qualitative variables were described according to absolute and relative frequency distributions. Quantitative variables were evaluated using central trend measures (mean and median) and scatter measures (s.d.). Both Pearson’s chi-square and Fisher's exact tests were performed for testing the null hypothesis of independence of variables in contingency tables.
Disease-free survival was defined as the time from diagnosis until disease progression or death by any cause on the date of the last follow-up. Overall survival was defined as the time from diagnosis until death by any cause. We used the Kaplan–Meier method to estimate disease-free survival and overall survival curves. The survival distributions for the different values of the patients’ characteristics were compared using the log-rank test. Cox proportional hazards models for disease-free survival and overall survival were also fitted, using the Efron approximation for tie handling and including patients’ characteristics.
Results
A total of 67 cases of male breast cancer with enough tumor tissue to be classified both with the validated six-marker immunohistochemical panel and PAM50 signature were analyzed retrospectively. The median age was 64 years (range: 23–92). The clinicopathological characteristics of these patients are described in Table 1. Thirty patients (45%) had T1 tumors and 29 (43%) positive lymph nodes. Only three patients (5%) presented advanced disease at diagnosis. The predominant histology was invasive carcinoma of no especial type (ductal, NOS) (90%). Sixty-four cases (96%) were ER-positive, 56 (84%) were PR-positive and 60 (90%) were AR-positive. Only three patients (5%) had a tumor that was ER and PR-negative. The only patient displaying a Her2-positive tumor by immunohistochemistry was confirmed to be positive by CISH and was treated with trastuzumab in the adjuvant setting.
All patients were treated surgically with either mastectomy in 57 patients (85%) or lumpectomy in 10 patients (15%). Thirty-seven (55%) patients underwent axillary dissection, 23 (34%) had sentinel node biopsy alone and 7 (11%) no axillary surgery. Thirty-seven percent of patients received adjuvant radiotherapy. Some patients also received other adjuvant systemic treatments, including hormonal therapy (85%) and or chemotherapy (55%). Twelve (18%) patients had a family history of breast cancer and only three (5%) had a personal history of gynecomastia. Five patients (7%) developed a second cancer after breast cancer history: 2 prostate cancers, 1 bladder cancer and 2 melanomas. No metachronous contralateral breast cancers were observed.
The distribution of tumor molecular subtypes according PAM50 signature was as follows: 40 (60%) luminal B, 20 (30%) luminal A, 7 (10%) Her2-enriched and none of the tumors were basal-like. Using immunohistochemical panel surrogates, 34 (51%) were luminal B, 29 (44%) luminal A, 2 (3%) non-basal triple-negative, 1 (1%) basal-like and 1 (1%) Her2-positive. Those triple-negative samples (basal and non-basal) by immunohistochemistry were 1 luminal A, 1 luminal B and 1 Her2-enriched by PAM50. We found a strong correlation between immunohistochemistry and PAM50 subtyping in our cohort (P=0.018) when comparing patients grouped as luminal A, luminal B, Her2-positive/Her2-enriched or triple-negative/basal-like by both classifications. The distribution of PAM50 subtypes within luminal A and luminal B defined by immunohistochemistry are displayed in Figure 1. As we can observe, despite the strong association between immunohistochemical and PAM50 subtyping, more than half of luminal A patients by immunohistochemistry are classified in a different group by PAM50. From the patients classified as Her2-enriched by PAM50, only one was Her2-positive by immunohistochemistry and CISH, being the rest negative by both immunohistochemistry and CISH. Of note, this Her2-enriched/Her2-positive patient was also ER and PR-positive, and could be considered as luminal B like (Her2-positive) depending on the classification criteria used.14 In order to verify if there were any Her2 subclones missed by the placement of small-sized cores into tissue microarrays, we assessed the Her2 status of the 6 Her2-negative/Her2-enriched tumor samples in their original tissue blocks and confirmed that all of them were Her2-negative.
There were no significant differences between the clinicopathological features of our cohort of PAM50 luminal A and luminal B tumors as shown in Table 2. There was only a trend toward a higher percentage of luminal B subtype tumors that were poorly differentiated (grade 3) (38% vs 10%, P=0.08) compared with luminal A tumors by PAM50.
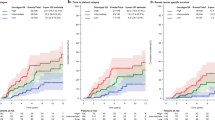
With a median follow-up of the patients of 128 months (range: 93–162 months), 17 (25%) had recurrence: 2 had local relapse and 15 distant metastases. Twenty patients (30%) died during follow-up, 12 (18%) from progression of the disease, and 8 (12%) from other causes unrelated to the tumor. We found no significant differences between luminal A and luminal B by PAM50 in disease-free survival (OR 0.29 (0.07–3.11), P=0.291) or overall survival (OR 0.46 (0.15–4.91), P=0.8). Similarly, there were no differences between luminal A and luminal B based on the immunohistochemical panel in disease-free survival or overall survival (data not shown). When we analyzed the differences in overall survival between Her2-enriched and non-Her2-enriched by PAM50 signature, we found a significant worse outcome in Her2-enriched tumors (median of 71 vs 128 months, respectively; OR 2.59 (0.47–14.29), P=0.046) (Figure 2).
Discussion
To our knowledge, this is the first study that classifies male breast cancer in molecular subtypes based on PAM50 signature and correlates them to immunohistochemical subtyping. Our results confirm that male breast cancer is mainly a genomic luminal disease with a strong correlation between PAM50 and a validated six-marker immunohistochemical panel, being the majority of the samples luminal B (60% by PAM50 and 51% by the immunohistochemical panel) followed by luminal A (30% and 44%, respectively). It is worth noting that only one Her2-enriched patient by PAM50 was also Her2-positive by immunohistochemistry and CISH. Therefore, he was the only one receiving trastuzumab. The rest of the PAM50 Her2-enriched patients did not receive any anti-Her2 therapy and had worse outcome compared with luminal tumors. We did not identify any basal-like tumors by the PAM50 assay.
We found several unique characteristics of male breast cancer compared with female breast cancer, including higher rates of ER positivity (96%), PR positivity (84%) and AR positivity (90%), lower Her2 positivity (1%), older age at presentation (median age 64 years) and higher proportion of nodal disease (43%), although a direct comparison between both types of tumors is lacking. These results were comparable to the data reported by the EORTC10085/TBCRC/BIG/NABCG International Male Breast Cancer Program retrospective analysis, the largest collection of male breast cancer clinical characteristics and biological samples to date (n=1483).15
Several research groups have attempted to classify male breast cancer into molecular subtypes using the same immunohistochemical classification of female breast cancer and the subsequent clinical outcome.16, 17, 18, 19, 20 These reports have in common that, the majority of the tumors were classified as luminal A (60–80%) defined as ER-positive and/or PR-positive and Her2-negative. Luminal B subtype defined as ER-positive and/or PR-positive and Her2-positive was the second more frequent subtype after luminal A. They found no correlation in prognosis between luminal A vs luminal B tumors in terms of breast cancer recurrence-free and disease-specific survival. Fewer triple-negative (basal-like) (0–6%) and Her2-positive (0–9%) tumors were reported compared to female breast cancer, and were associated with a worse prognosis (Table 3).
On the contrary, the studies of Kornegoor and Piscuoglio21, 22 like ours23 differentiated between luminal A and luminal B subtypes according to differences in the expression of Ki-67 as proliferation marker with a cut-off point of 14%. This definition was adopted by the St Gallen Expert Consensus Panel recommendation guidelines for the systemic treatment of early breast cancer and improved the distinction between luminal A and luminal B tumors.14 However, there are still many concerns about this classification method because of the considerable lack of reproducibility between laboratories, as methods are not yet standardized, and the optimal cut-off point of Ki-67 has not been well defined yet. Therefore, the use of immunohistochemical markers may result in the misclassification of many tumors compared with the information generated by gene expression profiling.
There is a growing consensus about multigene prognostic gene signatures providing useful complementary information to clinical and pathological data in ER-positive breast cancer. In contrast to female breast cancer, the genetic landscape of male breast cancer has yet to be fully characterized and there have only been a few array based studies investigating DNA copy number aberrations and gene expression profiles. Johansson et al24, 25 classified male breast cancer tumors into two molecular subgroups based on copy number alterations and gene expression profiling with differences in tumor biology features and outcome. The luminal M1 subgroup included more aggressive tumors with high level of chromosomal changes and had upregulated genes in cell proliferation, migration, tumor invasion and metastasis. The luminal M2 subgroup had less chromosomal alterations and had upregulated immune response genes and ER signaling-associated genes. Interestingly, they did not resemble any of the subgroups reported for female breast cancer. Although most male breast cancers are ER-positive, not all of them are similar and they may behave differently to ER-positive female breast cancers because of a gender associated landscape in hormone receptor pathways.
Male breast cancers may also differ from female breast cancers in their mutational status repertoire and the mutational frequency of the most commonly mutated genes. Piscuoglio et al22 reported that male breast cancer had less PIK3CA and TP53 mutations than female breast cancer of the same immunohistochemical profile but displayed more frequently mutations in genes associated with DNA repair genes. These findings suggest that it may relevant to investigate the potential use of therapeutic agents targeting DNA repair defects in male breast cancer. Furthermore, as an exploratory hypothesis-generating analysis, there were distinct repertoires of somatic mutations between luminal A and luminal B, being, for example, DNA repair genes more frequently mutated in luminal B (33%) than luminal A male breast tumors (6%).
PAM50 test classifies breast tumors into intrinsic subtypes (luminal A, luminal B, Her2-enriched and basal-like) and can be used to estimate the distant recurrence-free survival for postmenopausal women with hormone receptor positive, early-stage breast cancer to be treated with adjuvant endocrine therapy when combined with the Prosigna algorithm. We have used for the first time the PAM50 signature to unravel the molecular heterogeneity in male breast cancer. The majority of tumors in our cohort were classified as a genomic luminal disease in correlation with a validated six-marker immunohistochemical panel. The open question is why most male breast cancers are hormone receptor positive. Several hypotheses can be raised to explain this finding but the real explanation is still unknown. As with female breast cancer, the rate of hormone receptor positivity increases with age, with a mean age at diagnosis of breast cancer higher in males than in females. Although the majority of men with breast cancer have no identifiable risk factors, several have been associated with an imbalance in estrogen excess or lack of androgen (chronic liver diseases, cryptorchidism, Klinefelteŕs syndrome). Farhat et al,26 in a prospective study of healthy postmenopausal women who were not taking hormone therapy, observed that higher serum estradiol levels were associated with an increased risk of ER-positive breast cancer, although there is no information available in men. However, Callari et al27 reported a different pattern of gene expression of steroid receptors in male breast cancer specimens compared with ER-positive female breast cancer samples with similar clinicalopathological features, indicating that there may be many differences between the biology of male and female breast cancer.
In our cohort of male breast cancers, we found a higher prevalence of luminal B tumors characterized by a more aggressive biology, higher proliferation and less endocrine responsiveness in contrast with the higher frequency of the luminal A subtype in female breast cancer. It should be noted that, despite there was a strong association between immunohistochemical and PAM50 subtyping (P=0.018), we found some discordances between these classifications. More than 50% of luminal A patients by immunohistochemistry were classified in a different group by PAM50 although we do not know if this may have an impact in the treatment of male breast cancer patients. Nevertheless, we did not find significant differences in the clinicopathological features or in the outcome between both luminal A and luminal B subtypes. Most tumors were diagnosed in elderly patients, some of whom died from other causes unrelated to the tumor, and relapses can occur even a long time after the initial diagnosis, which may explain at least partially why we did not observe differences in disease-free survival and overall survival between luminal A and luminal B subtypes.
In the other hand, our patients with Her2-negative (immunohistochemistry)/Her2-enriched (PAM50) tumors were unlike to receive any anti-Her2 therapy and had a worse outcome compared with luminal tumors. The incorporation of anti-Her2 therapy changed beneficially the natural history of Her2-positive breast cancer from a historically aggressive disease tumor subtype, and also improved their outcome beyond of Her2-negative breast cancer.28 Data from the Cancer Genome Atlas breast cancer29 suggested that a subset of Her2-positive tumors with a Her2-enriched gene expression profile by PAM50 had the highest activation of the Her2/EGFR signaling pathway and may benefit the most from anti-Her2 therapies. Recently, several studies30, 31, 32 have provided clinical evidence suggesting that Her2-enriched tumors had the highest pCR compared Her2-positive or any other subtypes, and so that might benefit the most from Her2 targeting therapies. In the metastatic setting, Prat et al,33 in an unplanned retrospective analysis from the EGF30008 phase 3 clinical trial, reported a benefit from lapatinib therapy in a small group of patients with Her2-negative/Her2-enriched metastatic breast cancer with poor outcome.
Although the results from our study are limited by the number of patients analyzed and the study’s retrospective nature, we consider that it provides useful information on the biology of male breast cancer patients and their outcome over time. In contrast with female breast cancer, in which a third of the cases are non-luminal, our findings suggest that male breast cancer is mainly a genomic luminal disease based on PAM50 gene signature. More research is needed to identify the reason for the scarcity of non-luminal subtypes in male breast cancer, as well as the predominance of the luminal B followed by luminal A subtypes and its clinical significance. In addition, we found a proportion of patients with a Her2-negative/Her2-enriched profile untreated with any anti-Her2 therapy with a worse outcome compared with luminal subtypes. Identifying these patients and the subsequent treatment with anti-Her2 therapy could change the natural history of their disease.
References
Korde LA, Zujewski JA, Kamin L et al, Multidisciplinary meeting on male breast cancer: summary and research recommendations. J Clin Oncol 2010; 28: 2114–2122.
Giordano SH . A review of the diagnosis and management of male breast cancer. Oncologist 2005; 10: 471–479.
Perou CM, Sørlie T, Eisen MD et al, Molecular portraits of human breast tumors. Nature 2000; 406: 747–752.
Nielsen TO, Hsu FD, Jensen K et al, Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res 2004; 10: 5367–5374.
Cheang MC, Voduc D, Bajdik C et al, Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res 2008; 14: 1368–1376.
Cheang MC, Chia SK, Voduc D et al, Ki-67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst 2009; 101: 736–750.
Rivenbark AG, O'Connor SM, Coleman WB . Molecular and cellular heterogeneity in breast cancer: challenges for personalized medicine. Am J Pathol 2013; 183: 1113–1124.
Sørlie T, Perou CM, Tibshirani R et al, Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 2001; 98: 10869–10874.
Sørlie T, Tibshirani R, Parker J et al, Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA 2003; 100: 8418–8423.
Wallden B, Storhoff J, Nielsen T et al, Development and verification of the PAM50-based Prosigna breast cancer gene signature assay. BMC Med Genomics 2015; 8: 54.
Gnant M, Filipits M, Greil R et al, Predicting distant recurrence in receptor-positive breast cancer patients with limited clinicopathological risk: using the PAM50 risk of recurrence score in 1478 postmenopausal patients of the ABCSG-8 trial treated with adjuvant endocrine therapy alone. Ann Oncol 2014; 25: 339–345.
Hammond ME, Hayes DF, Dowsett M et al, American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. Arch Pathol Lab Med 2010; 134: e48–e72.
Wolff AC, Hammond ME, Hicks DG et al, Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol 2013; 31: 3997–4013.
Goldhirsch A, Winer EP, Coates AS et al, Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol 2013 24: 2206–2223.
Vermeulen MA, Slaets L, Cardoso F et al, Pathological characterisation of male breast cancer: results of the EORTC 10085/TBCRC/BIG/NABCG International Male Breast Cancer Program. Eur J Cancer 2017; 82: 219–227.
Ge Y, Sneige N, Eltorky MA et al, Immunohistochemical characterization of subtypes of male breast carcinoma. Breast Cancer Res 2009; 11: R28.
Shaaban AM, Ball GR, Brannan RA et al, A comparative biomarker study of 514 matched cases of male and female breast cancer reveals gender-specific biological differences. Breast Cancer Res Treat 2012; 133: 949–958.
Yu XF, Feng WL, Miao LL et al, The prognostic significance of molecular subtype for male breast cancer: a 10-year retrospective study. Breast 2013; 22: 824–827.
Nilsson C, Johansson I, Ahlin C et al, Molecular subtyping of male breast cancer using alternative definitions and its prognostic impact. Acta Oncol 2013; 52: 102–109.
Leone JP, Leone J, Zwenger AO et al, Prognostic significance of tumor subtypes in male breast cancer: a population-based study. Breast Cancer Res Treat 2015; 152: 601–609.
Kornegoor R, Verschuur-Maes AH, Buerger H et al, Molecular subtyping of male breast cancer by immunohistochemistry. Mod Pathol 2012; 25: 398–404.
Piscuoglio S, Ng CK, Murray MP et al, The genomic landscape of male breast cancers. Clin Cancer Res 2016; 22: 4045–4056.
Sánchez-Muñoz A, Román-Jobacho A, Alba E et al, Male breast cancer: immunohistochemical subtypes male breast cancer: immunohistochemical subtypes and clinical outcome characterization. Oncology 2012; 83: 228–233.
Johansson I, Nilsson C, Berglund P et al, High-resolution genomic profiling of male breast cancer reveals differences hidden behind the similarities with female breast cancer. Breast Cancer Res Treat 2011; 129: 747–760.
Johansson I, Nilsson C, Berglund P et al, Gene expression profiling of primary male breast cancers reveals two unique subgroups and identifies N-acetyltransferase-1 (NAT1) as a novel prognostic biomarker. Breast Cancer Res 2012; 14: R31.
Farhat GN, Cummings SR, Chlebowski RT et al, Sex hormone levels and risks of estrogen receptor-negative and estrogen receptor-positive breast cancers. J Natl Cancer Inst 2011; 103: 562–570.
Callari M, Cappelletti V, De Cecco L et al, Gene expression analysis reveals a different transcriptomic landscape in female and male breast cancer. Breast Cancer Res Treat 2011; 127: 601–610.
Dawood S, Broglio K, Buzdar AU et al, Prognosis of women with metastatic breast cancer by HER2 status and trastuzumab treatment: an institutional-based review. J Clin Oncol 2010; 28: 92–98.
Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumors. Nature 2012; 490: 61–70.
Carey LA, Berry DA, Cirrincione CT et al, Molecular heterogeneity and response to neoadjuvant human epidermal growth factor receptor 2 targeting in CALGB 40601, a randomized phase III trial of paclitaxel plus trastuzumab with or without lapatinib. J Clin Oncol 2016; 34: 542–549.
Prat A, Bianchini G, Thomas M et al, Research-based PAM50 subtype predictor identifies higher responses and improved survival outcomes in HER2-positive breast cancer in the NOAH study. Clin Cancer Res 2014; 20: 511–521.
Santonja A, Ribelles N, Jimenez-Rodriguez B et al, Abstract P3-07-14: Prosigna® intrinsic subtyping predicts response to neoadjuvant combination therapy in study that includes herceptin within HER2+ (IHC) patients [abstract]. Cancer Res 2016; 76 (4 Supp): Abstract nr P3-07-14; doi: 10.1158/1538-7445.SABCS15-P3-07-15.
Prat A, Cheang MC, Galván P et al, Prognostic value of intrinsic subtypes in hormone receptor-positive metastatic breast cancer treated with letrozole with or without lapatinib. JAMA Oncol 2016; 2: 1287–1294.
Acknowledgements
We gratefully acknowledge Nanostring Technologies team for providing the reagents for PAM50 subtypes determination as well as technical support, Maria José Lozano for her support with samples immunostaining and Jose M Roldan for the edition of the artwork. Angela Santonja has a predoctoral grant PFIS-ISCIII (FI12/00489).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Sánchez-Muñoz, A., Vicioso, L., Santonja, A. et al. Male breast cancer: correlation between immunohistochemical subtyping and PAM50 intrinsic subtypes, and the subsequent clinical outcomes. Mod Pathol 31, 299–306 (2018). https://doi.org/10.1038/modpathol.2017.129
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2017.129