Abstract
Endometrial cancer is the most common cancer of the female genital tract in developed countries. Although the majority of endometrial cancers are diagnosed at early stages and the 5-year overall survival is around 80%, early detection of these tumors is crucial to improve the survival of patients given that the advanced tumors are associated with a poor outcome. Furthermore, correct assessment of the pre-clinical diagnosis is decisive to guide the surgical treatment and management of the patient. In this sense, the potential of targeted genetic sequencing of uterine aspirates has been assessed as a pre-operative tool to obtain reliable information regarding the mutational profile of a given tumor, even in samples that are not histologically classifiable. A total of 83 paired samples were sequenced (uterine aspirates and hysterectomy specimens), including 62 endometrioid and non-endometrioid tumors, 10 cases of atypical hyperplasia and 11 non-cancerous endometrial disorders. Even though diagnosing endometrial cancer based exclusively on genetic alterations is currently unfeasible, mutations were mainly found in uterine aspirates from malignant disorders, suggesting its potential in the near future for supporting the standard histologic diagnosis. Moreover, this approach provides the first evidence of the high intra-tumor genetic heterogeneity associated with endometrial cancer, evident when multiple regions of tumors are analyzed from an individual hysterectomy. Notably, the genetic analysis of uterine aspirates captures this heterogeneity, solving the potential problem of incomplete genetic characterization when a single tumor biopsy is analyzed.
Similar content being viewed by others
Main
Endometrial cancer is the fourth most common cancer among women in developed countries and the most frequent cancer of the female genital tract.1 Endometrial cancer is mainly classified into two groups with different clinical, pathological, and molecular features.2, 3 Type I or endometrioid endometrial carcinomas are normally low-grade, estrogen-related tumors with a good prognosis. These tumors are the most common endometrial cancers and they usually arise in perimenopausal women, preceded by or coexisting with endometrial hyperplasia. Type II tumors are high-grade non-endometrioid endometrial carcinomas, unrelated to estrogen, which occur in older women and have a poor prognosis.4, 5 The majority of non-endometrioid endometrial carcinomas are serous endometrial carcinomas, although there are also less frequent histological subtypes, such as uterine carcinosarcomas (also known as malignant mixed müllerian tumors), uncommon biphasic neoplasms with malignant epithelial elements, and a sarcomatoid component.5 At the molecular level, significant differences are evident between type I and II carcinomas. Mutations in PTEN, PIK3CA, PIK3R1, KRAS, CTNNB1, FGFR2, and ARID1A are associated with endometrioid endometrial carcinomas, whereas mutations in TP53, PIK3CA, and PPP2R1A are frequent in non-endometrioid endometrial carcinomas.6, 7 In addition, microsatellite instability is evident in one-third of endometrioid endometrial carcinomas, a feature that is unusual in non-endometrioid endometrial carcinomas, which are more commonly characterized by chromosomal instability.8, 9
The majority of endometrial cancers are diagnosed at early stages and the associated 5-year overall survival is around 80%. Nevertheless, the survival rate decreases to 57–46% for high-grade tumors.5 Furthermore, carcinosarcomas account for a high percentage of mortality despite constituting only 5–6% of endometrial cancers, principally because 60% of the patients presents extra uterine disease at the moment of diagnosis. In these cases more than 50% will suffer recurrence after surgery and adjuvant treatment.10, 11
As with many other tumors, early detection of endometrial cancer is crucial to increase patient survival, particularly as advanced tumors are associated with a worse outcome. Moreover, the correct assessment of pre-clinical diagnosis is also decisive, as this will guide the pre-operative and surgical management of the patient.12 In this sense, the use of uterine aspirates (Pipelle biopsies) as diagnostic pre-operative biopsies is widely recommended, representing a minimally invasive and highly sensitive procedure. However, the failure rate in obtaining such samples is around 8%, whereas 13% of the samples turn out to be histologically inadequate, figures that are significantly higher in postmenopausal women.12, 13 Moreover, discrepancies between pre- and post-operative biopsies have been observed with respect to histological grade, which could lead to a misclassification and the use of inappropriate therapeutic strategies.14, 15 As such, there has been some interest in identifying molecular markers in uterine aspirates, enhancing their potential as a diagnostic sample for both histological classification and molecular characterization of tumors.16, 17, 18
Describing the genetic profile of tumors can be decisive for their accurate diagnosis and for therapeutic decision-making. However, intra-tumor genetic heterogeneity represents a challenge that hampers the correct characterization of tumor samples.19 The current study reveals how uterine aspirates are a potentially useful tool to circumvent the problems derived from intra-tumor heterogeneity when genetically characterizing endometrial cancer. We defined the mutational profile of endometrial cancers in paired pre-operative uterine aspirates and hysterectomy specimens from patients. The data obtained not only confirmed the utility of these aspirates to detect the mutations in primary tumors, even when a pathological diagnosis could not be achieved by other means, but importantly, they also reflected the high intra-tumor genetic heterogeneity found in endometrial cancers. These results show that the genetic analysis of uterine aspirates provides information that the pathologist may find useful to reduce the rate of false-negative diagnoses. In summary, we show the importance of uterine aspirates in studying endometrial cancer at the molecular level, supporting the potential of non-invasive biopsies for the diagnosis and characterization of certain tumor types.
Materials and methods
Sample Description
A total of 62 endometrial cancer cases (44 endometrioid endometrial carcinomas, 9 serous endometrial carcinomas, 9 carcinosarcomas) were collected at Vall d’Hebron Hospital (Barcelona), Arnau de Vilanova University Hospital (Lleida), MD Anderson Cancer Center (Madrid) and Medical University (Lubin) between 2010 and 2015. The median age of the patients was 67 (±12, endometrioid endometrial carcinomas), 75 (±8, serous endometrial carcinomas), and 72 (±8, carcinosarcomas) and the histopathological data of the tumors studied can be found in Supplementary Table 1. Endometrial tissue from endometrial aspirates and hysterectomy specimens were analyzed from each subject. In addition, samples obtained from 10 patients diagnosed with atypical hyperplasia, were collected at Hospital Universitari de Bellvitge and used as an example of precursor malignant neoplasia. A total of 27 patients not diagnosed with cancer were also analyzed as controls for the studies of the mutational profile (7 non-atypical hyperplasia endometrium, the endometrium from 7 patients with leiomyoma, and 13 normal endometrium). In 11 of these controls uterine aspirates and their respective hysterectomy specimen (endometrial tissue) were analyzed, whereas in the remainder only a uterine aspirate was available. Uterine aspirates were collected using a Pipelle de Cornier to obtain the sample that was then centrifuged for 20 min, as described previously.16 The pellet containing the cells from the uterine cavity was processed as formalin-fixed and paraffin-embedded tissue for further DNA extraction. A second uterine aspirate was obtained in the operating room just before surgery, being the tumor material frozen at −80 °C for hematoxylin and eosin stain examination and DNA extraction. Only in which the formalin-fixed and paraffin-embedded uterine aspirate material was used up in the histologic analysis, frozen tissue was used for the study. The study was approved by the local ethical committee from each institution, and a complete written informed consent was obtained from all patients.
DNA Extraction and Mutational Analysis
DNA was obtained from formalin-fixed paraffin-embedded and frozen samples using phenol extraction and ethanol precipitation, and 10 ng were used for sequencing. Multiplex PCR to prepare amplicon libraries was performed using the Ion AmpliSeq Library Kit 2.0 and Ion AmpliSeq Cancer Hotspot Panel v2 (Life Technologies). For PCR, a total of 17 and 20 cycles were used for the frozen and formalin-fixed paraffin-embedded samples, respectively. The PCR template preparation and enrichment were performed using Ion PGM Template OT2 200 Kit and the Ion OneTouch 2 System. Finally, the Ion PGM Sequencing 200 Kit v2 and Ion PGM System (Life Technologies) were used for DNA sequencing according to the manufacturer’s protocols. Duplicates were analyzed for 10 % of the samples, rendering equivalent results. For the bioinformatics analysis, see Supplementary Methods.
Sanger Sequencing
To validate the mutations, a total of 88 of the 476 variants found in the samples analyzed were Sanger sequenced. The PCR conditions and amplicon lengths used are indicated in Supplementary Table 2. Only 6 of these variants were not confirmed by Sanger sequencing, which was probably due to their poor quality and/or their frequencies below 10% in the Ion PGM sequencing analysis (Supplementary Table 3).
Statistical Analysis
A paired t-test was used to compare the data from the hysterectomy tumor samples and uterine aspirates. Two-tailed tests were performed and 95% confidence intervals (CIs) were accepted. The mutation discovery rate was calculated in each sample (aspirate or tumor region) from the same patient according to the following equation:

The Pearson coefficient was used to analyze the correlation between the percentage of tumor cells in patient samples and the MDR. P values <0.05 were considered statistically significant and the statistical analyses were performed using the SPSS Statistics 17.0 software (SPSS, Chicago, IL, USA).
Results
Identification of the Mutational Profile in Paired Uterine Aspirate and Hysterectomy Specimen Samples
Uterine aspirates are thought to be highly sensitive and specific biopsies for the pre-operative diagnosis of endometrial cancer, especially when based on biomarker expression.13, 16, 17, 18 To investigate the usefulness of mutation detection in uterine aspirates, the molecular profile of paired samples (pre-operative uterine aspirates and the corresponding resected surgical specimen) from 54 patients with endometrial cancer (37 endometrioid endometrial carcinomas, 9 serous endometrial carcinomas, and 8 carcinosarcomas) and 10 patients with atypical hyperplasia was analyzed using AmpliSeq Cancer Hotspot Panel v2. This panel analyzes approximately 2800 cancer mutations of 50 oncogenes and tumor suppressor genes, some of which are frequently altered in endometrial cancer (PTEN, KRAS, FGFR2, CTNNB1, PIK3CA, FBXW7, and TP53). In addition, a total of 27 patients not diagnosed with cancer were also analyzed as control cases (7 cases of non-atypical hyperplasia, 7 cases with leiomyomas and 13 with a normal endometrium).
Sequencing analysis revealed the presence of mutations in 51 of the 54 aspirates from cancer patients (Table 1; Supplementary Table 3A) and in 5 of the 10 aspirates from atypical hyperplasia cases (Supplementary Table 4). By contrast, mutations were only identified in 1 of the 27 control patients (data not shown). Although it is currently unfeasible to reach a diagnosis of endometrial malignancies based exclusively on genetic alterations, these results indicate that genetic analysis of uterine aspirates may offer reliable support to histological diagnosis.
Mutations identified in the different subgroups of patients were consistent with previous studies.6, 7 In summary, endometrioid endometrial carcinomas carried mutations in PTEN (71.1% of patients), PIK3CA (39.5%), CTNNB1 (28.9%), TP53 (28.9%), FGFR2 (23.7%), KRAS (21.1%), and CDKN2A (10.5%). In addition, we also detected mutations in less commonly affected genes, such as: ABL1, AKT1, APC, ATM, BRAF, ERBB2, FBXW7, KIT, RB1, and VHL1 (5.3%); and GNA11, GNAS, HNF1A, MET, MLH1, NRAS, RET, STK11, SMAD4, SMARCB1, and SMO (2.6%). As expected, the most frequently mutated gene in serous carcinomas and carcinosarcomas samples was TP53 (77.7 and 87.5%, respectively). Frequencies found in our series were generally higher than those detected in the The Cancer Genome Atlas dataset6 (Supplementary Figure 1A). This could be explained taking into account the sequencing method applied in each study. Whereas The Cancer Genome Atlas study6 performed whole-exome sequencing (mean coverage around to 50 ×) our study has been developed with targeted sequencing (mean coverage around to 1000 ×), allowing to detect more accurately the mutations, specially those with low frequency.20 TP53 mutation frequency was particularly high in our series, probably due to the presence of mutations in 8 of the 24 high-grade endometrioid carcinomas (Supplementary Figure 1B). To be sure that these cases were not misclassified a second pathology review was performed, confirming the initial diagnosis (Supplementary Table 5).
To gain further insight into the suitability of uterine aspirates to detect mutations and consequently, to estimate the potential of uterine aspirates to characterize endometrial caner from a genetic point of view, we analyzed the percentage of pathogenic variants present in hysterectomy specimens that were also detected in aspirates (Figure 1). All the mutations detected in the surgical tumor tissue were also found in 30 out of the 36 aspirates (83.3%) from endometrioid endometrial carcinoma patients. In terms of the rest of the samples, 50–75% of the mutations detected in the hysterectomy specimen also appeared in the corresponding aspirate in three of them (8.3% of the total), although in two of them (5.6% of the total) the aspirate contained 25–50% of the mutations present in the surgical tissue. Only in 1 patient did we fail to detect any of the mutations identified in the hysterectomy sample in the corresponding aspirate, accounting for 2.8% of the total cases. Conversely, in 1 other patient mutations were detected in the uterine aspirate, whereas none were identified in the surgical sample (Table 1). Furthermore, in 6 of the 9 uterine aspirates from serous carcinomas patients 100% of the mutations identified in the hysterectomy specimens were detected in the aspirate (66.7% of the total cases), as in 6 of the 8 carcinosarcoma cases (75% of the total).
Percentage of mutations in hysterectomy specimens identified in paired uterine aspirate. Graphs represents the percentage (100%, 75–50%, 50–25%, or 0%) of the mutations found in surgical tumor samples and paired aspirates in endometrioid carcinoma (a), serous carcinoma (b), and carcinosarcoma (c) samples.
Misclassifying the histological grade of pre-operative biopsies can have grave consequences14, 15, 18 and indeed, in our samples 22 of the 37 endometrioid endometrial carcinomas uterine aspirates (59.5%) were misclassified with respect to their grade during the pathological diagnosis, the majority of them being attributed with a lower grade than that detected in the definitive hysterectomy specimen (Table 1). However, in 17 of these 22 (77.2%) discordant classifications, the uterine aspirates were concordant in the mutational analysis, showing all the mutations detected in their respective surgical specimen. Nevertheless, no relationship between mutational status and histological type or grade has been previously described, and nor was one found in our series. Consequently, these results confirm that the genetic analysis of uterine aspirates as a pre-operative biopsy can reliably reproduce the molecular status of the tumor in a pre-clinical setting. However, further studies into the mutational profile and histological grade will be necessary to take the mutational information from uterine aspirates into account when assessing the tumor grade.
Genetic Analysis Helps to Reduce the Rate of False-Negative Diagnoses in Uterine Aspirates
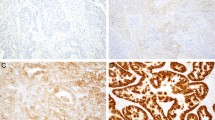
The histologic analyses of uterine aspirates fail to distinguish the presence or absence of malignancy in around 13% of the cases, either due to the small proportion of representative tumor cells or to the poor quality of the specimen.21, 22, 23 To further investigate the potential of genetic analysis of uterine aspirates as an informative tool for endometrial cancer diagnosis, we assessed the tumor mutations that could be detected in uterine aspirates that could not be evaluated on a pathological basis (Figure 2a). Mutational analysis was performed on eight paired samples of non-diagnosable uterine aspirates from patients who turned out to have endometrial cancer and on the corresponding hysterectomy specimens (7 endometrioid endometrial carcinomas and 1 carcinosarcoma). Interestingly, seven of the eight non-evaluable uterine aspirates had a similar mutation profile to that of their paired surgical sample (Figure 2b; Supplementary Table 3B). We did not find mutations in the uterine aspirate from one patient, as was also the case in the paired hysterectomy tumor tissue. We validated these results by Sanger sequencing and immunohistochemistry when material was available. For example, the CTNNB1 mutation in case EEC-38 was validated by Sanger sequencing in the aspirate and surgical tissue (Figure 2c). To validate this, we also analyzed β-catenin expression by immunohistochemistry in surgical tissue (Figure 2d). These results demonstrated that genetic sequencing complements pathological analysis and contributes significantly to a more comprehensive characterization of the tumor at very early stages of diagnosis, providing valuable information for its correct classification.
Genetic analysis of non-evaluable uterine aspirates. Paired samples of non-evaluable uterine aspirates and hysterectomy specimens were analyzed genetically. (a) Representative hematoxylin and eosin image of a uterine aspirate (upper image) and its paired surgical sample (lower image). (b) Summary of the mutations detected in the paired uterine aspirate and hysterectomy samples. Analysis of CTNNB1 (β-catenin) mutation (S37P) found in patient EEC-38 by (c) the Sanger sequencing in hysterectomy specimen and uterine aspirate samples and by (d) immunohistochemistry analysis. The white arrow label the nuclear localization of β-catenin, which is suggestive of mutations (panel magnification × 20).
Genetic Analysis of Uterine Aspirates Captures the Intra-Tumor Heterogeneity Found in Endometrial Carcinomas
It is well known that human cancers display substantial intra-tumor heterogeneity, not only in cellular morphology or gene expression but also in terms of genetic variation.24, 25 This phenomenon represents an important challenge to accurate diagnosis and therapeutic decision-making.19 Although recent studies showed intra-tumor genetic heterogeneity in gynecological cancers like ovarian cancer,26, 27 heterogeneity at the mutational level has not been described in endometrial cancer to our knowledge. Interestingly, the comparison between the mutational profile of uterine aspirates and hysterectomy specimens highlighted the presence of additional mutations in 11 out of the 54 uterine aspirates, mutations that were not present in the corresponding paired surgical tissue (Table 1). Thus, we examined whether these differences might reflect the intra-tumor genetic heterogeneity in this clinical context.
To explore this hypothesis, genetic sequencing analysis was performed on additional tumor regions from 21 of the endometrial cancer hysterectomy specimens previously studied (14 endometrioid endometrial carcinomas, 5 serous endometrial carcinomas and 5 carcinosarcomas: Table 2). Comparative mutation analysis revealed differences in the mutational profiles of the distinct regions of the endometrioid endometrial carcinomas tumor tissue analyzed from 10 out of 14 patients (71.4%), confirming the presence of intra-tumor heterogeneity (Supplementary Figure 2; Supplementary Table 3C). For example, in the three different tumor regions analyzed from case EEC-1 (Figure 3a), a total of 5 mutations in PTEN, TP53, and APC were detected, with one of the regions (tumor region 1) carrying all five, whereas the other two (tumor region 2 and 3) carried 2 and 3 mutations, respectively. In the remaining cases (4/14, 28.6%), a similar mutational profile was seen in all the samples analyzed (Supplementary Figure 2; Supplementary Table 3C), suggesting that these cases did not harbor significant intra-tumor heterogeneity, at least with respect to the genes and tumor regions studied. For example, this was the case of patient EEC-11 from whom all the samples analyzed carried mutations in FGFR2 and FBXW7 (Figure 3b). In contrast to the endometrioid endometrial carcinomas, intra-tumor heterogeneity was only detected in 1 of the 5 (20%) serous carcinomas and 1 of the 5 (20%) carcinosarcomas when additional tumor regions were analyzed (Supplementary Figure 3). The low proportion of mutational heterogeneity in cases with serous and carcinosarcoma histology could be due to the fact that chromosomal instability is a more frequent molecular alteration than punctual genetic changes in these tumor types,9 a modification that cannot be properly detected with the sequencing platform used here.
Characterization of the intra-tumor genetic heterogeneity in endometrial tumors. Representative mutational profile of genetically heterogeneous endometrioid carcinoma (a, EEC-1) and of a homogeneous endometrioid tumor (b, EEC-11). The colors in the squares represent the mutant allelic frequencies (MAFs). The squares marked as LQ identify low quality variants in the ion PGM analysis. The mutation discovery rate is defined as the percentage of mutations detected in each sample with respect to the totality of the mutations observed in all the samples analyzed from the same patient (see ‘Materials and methods’ section). The graph represents the mean mutation discovery rate (c) in endometrioid carcinomas, serous carcinomas, and carcinosarcomas. (*0.005<P<0.05; **0.001<P<0.005).
The sensitivity of mutation detection in each sample was scored as the mutation discovery rate, which indicates the proportion of mutations detected in each sample with respect to the total mutations observed in all the samples studied from a given patient (see ‘Materials and methods’ section). The mutation discovery rate was significantly higher in the endometrioid uterine aspirates than in the matched surgical tumor tissue, with a mean of 94.1% for uterine aspirates and 77.2% for individual hysterectomy tissue samples. This difference increased when low-quality mutations were not considered, decreasing the mutation discovery rate for surgical tumor samples to 67.5%, whereas the mutation discovery rate of the aspirates remained unaltered (Figure 3c). However, no significant differences were found in the serous carcinoma or carcinosarcoma samples. Differences in the mutation discovery rate are mainly found in heterogeneous tumors, due to the differences observed in the mutational profile between each tumor region (Figure 4). In 8 of the 10 (80%) heterogeneous endometrioid tumors, uterine aspirates reflected a higher mutation discovery rate than the tumor region used for the pathological diagnosis (tumor region 1). Only in one patient (EEC-7) the mutation discovery rate of the uterine aspirate was lower than that for the diagnostic tumor region, although it was equal or higher than that derived from the two other regions from that patient. In fact, the mutation discovery rate value was higher in uterine aspirates than in at least one tumor region in all cases where there was tumor heterogeneity. These differences seem not to be related to the proportion of the tumor tissue in each region analyzed as there was no significant correlation in a Pearson test (data not shown). These data confirm that genetic analysis of uterine aspirates detects a more representative mutational landscape of the tumor, reproducing in a single sample the intra-tumor heterogeneity found in the different tumor regions.
Mutation discovery rate in heterogeneous endometrioid endometrial carcinoma. The mutation discovery rate was calculated for each sample from the heterogeneous endometrioid carcinoma patients as indicated in the ‘Materials and methods’ section. Each bar represents a sample, from the bottom to the top: uterine aspirate and the different tumor regions.1, 2, 3, 4 The dark gray color represents the percentage of high quality variants detected and the light gray reflects the LQ variants identified in the ion PGM analysis.
Discussion
Advances in next-generation sequencing have revealed that genetic heterogeneity must be taken into account to fully understand tumor biology.19, 28, 29 Indeed, over and above the inter-patient heterogeneity,30 intra-tumor heterogeneity represents a real challenge for the precise characterization and adequate management of tumors.19, 31 The presence of different cell populations within a tumor with specific genomic, genetic and/or epigenetic characteristics has been demonstrated in numerous tumor types, including solid tumors and hematologic malignancies.19 Indeed, intra-tumor heterogeneity has been observed among gynecological cancers, particularly in high-grade serous ovarian carcinomas,27, 32, 33, 34 although this issue has not been studied in endometrial cancer so far. Therefore, a better understanding of the genetic heterogeneity underlying the biological and phenotypic evolution of endometrial is crucial to understand the clinical behavior of this disease. In this sense, the majority of the endometrioid carcinomas analyzed here have variable mutational profiles in the different tumor regions. By contrast, only 20% of serous carcinomas and 20% of carcinosarcomas showed mutational heterogeneity, which perhaps reflects the more frequent genetic mutations in endometrioid than in serous and carcinosacomas,6 the latter more often displaying large genomic changes.9 Therefore, a genomic study should be carried out on these tumor types to define the implication of copy number variation in intra-tumor heterogeneity, as previously described in high-grade serous ovarian carcinomas.27, 33
Intra-tumor clonal heterogeneity is thought to influ;ence therapeutic resistance and tumor progression,35 with some studies suggesting that some clones are genetically predisposed to resist therapy.36 In this context, characterizing intra-tumor heterogeneity would seem to be necessary to better predict the clinical outcome of a specific tumor at the moment of diagnosis and to establish the most appropriate treatment. The standard treatment for endometrial cancer is well established, involving surgery followed by adjuvant radiotherapy in tumors with a high-risk of recurrence. Chemotherapy is usually restricted to metastatic/recurrent and high-grade endometrial cancers, although traditional chemotherapy regimens are less effective than in cancers of other organs.5 In this sense, numerous clinical trials have been stratified according to genetic features, based on PTEN, PIK3CA, or FGFR3 mutational status. Consequently, tumor heterogeneity represents a therapeutic challenge and the use of a single diagnostic biopsy of a tumor may be insufficient, leading to the misclassification of a significant proportion of patients.
Several studies have centered on the feasibility of using liquid biopsies to analyze intra-tumor genetic heterogeneity.37, 38, 39, 40 In endometrial cancer, uterine aspirates are used as minimally invasive and highly sensitive biopsies for histological diagnosis or molecular characterization.16, 17, 18 In this regard, we found that the genetic analysis of uterine aspirates coupled to their pathological classification could be a very sensitive approach to detect endometrial malignant neoplasia. This implies that detecting a cancer-related mutation (such as those detected by the method we employed) is related to a possible malignant disorder or tumor. Although this seems to be true in our series it remains controversial, and a significantly larger number of samples (both normal and malignant) should be analyzed to address this issue. Paired sequencing of uterine aspirates and hysterectomy specimens confirms the efficacy in revealing malignant disorders (endometrial tumors or atypical hyperplasia) in uterine aspirates. Only three samples (5.5%) of uterine aspirates from tumor cases did not show any of the surgical tumor sample mutations, whereas a total of 42 (77.8%) of the aspirates carried all the mutations found in the corresponding hysterectomy specimen.
Furthermore, we detected mutations in aspirates that could not be evaluated pathologically. The amount of tissue obtained from endometrial biopsies from postmenopausal patients is sometimes insufficient to obtain an adequate diagnosis, which in the majority of cases is due to the presence of endometrial atrophy. However, patients with endometrial cancer on occasions provided poor quality samples. In a recent study of 1120 endometrial samples classified as unsuitable for diagnosis, a second biopsy was obtained from 38% of the patients that was suitable for diagnosis in 75% of cases, with 10% having a malignant tumor.23 Our results show that mutation analysis could indicate the presence of endometrial cancer or at least some pre-malignant anomaly, emphasizing the need for resampling in such cases and providing valuable information to accelerate the diagnosis.
Genetic analysis of uterine aspirates captures the intra-tumor heterogeneity identified in endometrioid endometrial carcinomas. The mutation discovery rate, defined as the percentage of mutations detected in each individual sample with respect to all the mutations found in a given patient, was used to measure the sensitivity of mutation detection in each sample. In heterogeneous tumors, the uterine aspirate mutation discovery rate was higher than that in at least one of the tumor regions. In fact, the mutation discovery rate value was higher in the uterine aspirate than in the tumor regions used for pathological diagnosis (tumor region 1) in 8 of the 10 heterogeneous endometrioid carcinomas. These results highlight the potential utility of this type of biopsy and reveal that the use of a unique tumor sample in diagnosis could underestimate the mutational burden in heterogeneous tumors. However, the study of multiple samples of a given tumor as a routine practice is still a difficult issue, as it would increase significantly the time and cost of diagnosis. Moreover, combining DNA from different tumor samples previously to the targeted sequencing is not a good option, because it would lead to a decrease in the frequency of those mutations, which are not present in all the tumor regions, causing some low-requency variants to be undetected. It is also worth pointing that it is fairly difficult to calculate how many tumor regions need to be analyzed to cover the intra-tumor heterogeneity found in each case. Taken together, these arguments increase the value of uterine aspirates as a genetic diagnostic biopsy, solving, at least in part, some of the problems found in the study of hysterectomy specimens. The fact that intra-tumor heterogeneity may be represented in uterine aspirates is probably related to the nature of such samples, consisting of cells from many different parts of the uterine cavity, which could provide a more representative picture of the entire tumor specimen than samples from a specific tumor region. Similar results were observed in ovarian carcinomas where intra-tumor genetic heterogeneity was evident when solid tumor biopsies were compared,32, 33, 34 but not when different ascites from the same patient were compared.41 In this case, ascites could represent the entire cavity in a similar way that uterine aspirates do in uterine cancers, capturing all the genetic mutations and representing the heterogeneity found in the solid tumor biopsies.
The use of non-invasive biopsies to diagnose and characterize tumors is currently a relevant clinical challenge. The data presented here shed light on the molecular characterization of minimally-invasive biopsies in endometrial cancer, and they provide potential solutions to the problem of detecting genetic heterogeneity, as well as valuable information in the case of biopsies with insufficient material. These data pave the way for the use of such analyses for other diseases.
References
Ferlay J, Soerjomataram I, Dikshit R et al.Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359–E386.
Bokhman JV . Two pathogenetic types of endometrial carcinoma. Gynecol Oncol 1983;15:10–17.
Lax SF, Kurman RJ . A dualistic model for endometrial carcinogenesis based on immunohistochemical and molecular genetic analyses. Verh Dtsch Ges Pathol 1997;81:228–232.
Llaurado M, Ruiz A, Majem B, et al. Molecular bases of endometrial cancer: new roles for new actors in the diagnosis and the therapy of the disease. Mol Cell Endocrinol 2012;358:244–255.
Yeramian A, Moreno-Bueno G, Dolcet X, et al. Endometrial carcinoma: molecular alterations involved in tumor development and progression. Oncogene 2013;32:403–413.
Cancer Genome Atlas Research Network Cancer Genome Atlas Research Network Kandoth C Cancer Genome Atlas Research Network Schultz N, et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013;497:67–73.
Hong B, Le Gallo M, Bell DW . The mutational landscape of endometrial cancer. Curr Opin Genet Dev 2015;30C:25–31.
Zighelboim I, Goodfellow PJ, Gao F, et al. Microsatellite instability and epigenetic inactivation of MLH1 and outcome of patients with endometrial carcinomas of the endometrioid type. J Clin Oncol 2007;25:2042–2048.
Tritz D, Pieretti M, Turner S, et al. Loss of heterozygosity in usual and special variant carcinomas of the endometrium. Hum Pathol 1997;28:607–612.
Cantrell LA, Blank SV, Duska LR . Uterine carcinosarcoma: a review of the literature. Gynecol Oncol 2015;137:581–588.
Artioli G, Wabersich J, Ludwig K, et al. Rare uterine cancer: carcinosarcomas. Review from histology to treatment. Crit Rev Oncol Hematol 2015;94:98–104.
Colombo N, Preti E, Landoni F, et al. Endometrial cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2013;24:vi33–vi38.
Clark TJ, Mann CH, Shah N, et al. Accuracy of outpatient endometrial biopsy in the diagnosis of endometrial cancer: a systematic quantitative review. BJOG 2002;109:313–321.
Karateke A, Tug N, Cam C, et al. Discrepancy of pre- and postoperative grades of patients with endometrial carcinoma. Eur J Gynaecol Oncol 2011;32:283–285.
Wang XY, Pan ZM, Chen XD, et al. Accuracy of tumor grade by preoperative curettage and associated clinicopathologic factors in clinical stage I endometriod adenocarcinoma. Chin Med J 2009;122:1843–1846.
Colas E, Perez C, Cabrera S et al.Molecular markers of endometrial carcinoma detected in uterine aspirates. Int J Cancer 2011;129:2435–2444.
Perez-Sanchez C, Colas E, Cabrera S, et al. Molecular diagnosis of endometrial cancer from uterine aspirates. Int J Cancer 2013;133:2383–2391.
Stelloo E, Nout RA, Naves LC, et al. High concordance of molecular tumor alterations between pre-operative curettage and hysterectomy specimens in patients with endometrial carcinoma. Gynecol Oncol 2014;133:197–204.
Jamal-Hanjani M, Quezada SA, Larkin J, et al. Translational implications of tumor heterogeneity. Clin Cancer Res 2015;21:1258–1266.
Sims D, Sudbery I, Ilott NE, et al. Sequencing depth and coverage: key considerations in genomic analyses. Nat Rev Genet 2014;15:121–132.
McCluggage WG . My approach to the interpretation of endometrial biopsies and curettings. J Clin Pathol 2006;59:801–812.
Phillips V, McCluggage WG . Results of a questionnaire regarding criteria for adequacy of endometrial biopsies. J Clin Pathol 2005;58:417–419.
Kandil D, Yang X, Stockl T et al.Clinical outcomes of patients with insufficient sample from endometrial biopsy or curettage. Int J Gynecol Pathol 2014;33:500–506.
Durrett R, Foo J, Leder K, et al. Intratumor heterogeneity in evolutionary models of tumor progression. Genetics 2011;188:461–477.
Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883–892.
Khalique L, Ayhan A, Whittaker JC, et al. The clonal evolution of metastases from primary serous epithelial ovarian cancers. Int J Cancer 2009;124:1579–1586.
Cooke SL, Ng CK, Melnyk N, et al. Genomic analysis of genetic heterogeneity and evolution in high-grade serous ovarian carcinoma. Oncogene 2010;29:4905–4913.
Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014;505:495–501.
Alexandrov LB, Nik-Zainal S, Wedge DC et al.Signatures of mutational processes in human cancer. Nature 2013;500:415–421.
Vogelstein B, Papadopoulos N, Velculescu VE et al.Cancer genome landscapes. Science 2013;339:1546–1558.
Kleppe M, Levine RL . Tumor heterogeneity confounds and illuminates: assessing the implications. Nat Med 2014;20:342–344.
Khalique L, Ayhan A, Weale ME et al.Genetic intra-tumour heterogeneity in epithelial ovarian cancer and its implications for molecular diagnosis of tumours. J Pathol 2007;211:286–295.
Bashashati A, Ha G, Tone A, et al. Distinct evolutionary trajectories of primary high-grade serous ovarian cancers revealed through spatial mutational profiling. J Pathol 2013;231:21–34.
Mota A, Trivino JC, Rojo-Sebastian A, et al. Intra-tumor heterogeneity in TP53 null high grade serous ovarian carcinoma progression. BMC Cancer 2015;15:940.
Pribluda A, de la Cruz CC, Jackson EL . Intratumoral heterogeneity: from diversity comes resistance. Clin Cancer Res 2015;21:2916–2923.
Bhang HE, Ruddy DA, Krishnamurthy Radhakrishna V, et al. Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nat Med 2015;21:440–448.
Patel KM, Tsui DW . The translational potential of circulating tumour DNA in oncology. Clin Biochem 2015;48:957–961.
De Mattos-Arruda L, Weigelt B, Cortes J, et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann Oncol 2014;25:1729–1735.
Raimondi C, Nicolazzo C, Gradilone A, et al. Circulating tumor cells: exploring intratumor heterogeneity of colorectal cancer. Cancer Biol Ther 2014;15:496–503.
Broersen LH, van Pelt GW, Tollenaar RA, et al. Clinical application of circulating tumor cells in breast cancer. Cell Oncol 2014;37:9–15.
Castellarin M, Milne K, Zeng T, et al. Clonal evolution of high-grade serous ovarian carcinoma from primary to recurrent disease. J Pathol 2013;229:515–524.
Acknowledgements
We thank all those at the Translational Research Laboratory and Immunohistochemical Laboratory from MD Anderson Madrid for their invaluable help. Tissue samples were obtained with the support of MD Anderson Foundation Biobank (Record Number B.0000745, ISCIII National Biobank Record), the ‘Xarxa Catalana de Bancs de Tumors’ and ‘Plataforma de Biobancos’ ISCIII (PT13/0010/0014, B.000609). This work was supported by grants from the AECC (Grupos Estables de Investigacion 2011-AECC- GCB 110333 REVE), the ‘Fundació La Marató, TV3’ (2/C/2013) to AG-M, JR, XM-G, and GM-B; Instituto de Salud Carlos III (ISCIII) (PI13/00132 and RETIC-RD12/0036/0007 to GM-B; RETIC-RD12/0036/0035 to JR; PI13/01701, and RD12/0036/0013 to XM-G; PI14/02043 and PI14/01942 to AG and MA); the ‘CIRIT, Generalitat de Catalunya’ (2014 SGR 1330 to JR; and 2014 SGR 138 to XM-G), GEIS award 2013 to GM-B and PG-S, and the ‘Communidad de Madrid’ (S2010/BMD-2303) to GM-B. AM is funded by the Spanish Ministry of Education, Culture, and Sports (FPU2012-5338). IC and PG-S are funded by PhD and postdoctoral contracts, respectively, from the AECC Scientific Foundation. EC is funded by the Spanish Ministry of Economy and Competitiveness (FPDI-2013-18322).
Author contributions
AM and PG-S performed the sequencing experiment and analysis. AM, EC, PG-S, and IC contributed to the sample processing. AR-S, SG, BD-F, AV, and AG performed the pathological analysis of the samples. LC, SA, AG-M, XG-T, PZ-M, and MB helped obtain the samples. MA and RL-L read and corrected the manuscript. EC, XM-G, JR, and GM-B conceived the study, participated in its design, and helped draft the manuscript. GM-B discussed and directed the study. All the authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Rights and permissions
About this article
Cite this article
Mota, A., Colás, E., García-Sanz, P. et al. Genetic analysis of uterine aspirates improves the diagnostic value and captures the intra-tumor heterogeneity of endometrial cancers. Mod Pathol 30, 134–145 (2017). https://doi.org/10.1038/modpathol.2016.143
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2016.143
This article is cited by
-
Cost-effectiveness analysis of molecular testing in minimally invasive samples to detect endometrial cancer in women with postmenopausal bleeding
British Journal of Cancer (2023)
-
Evolution of intra-tumoral heterogeneity across different pathological stages in papillary thyroid carcinoma
Cancer Cell International (2022)
-
Intratumor genetic heterogeneity and clonal evolution to decode endometrial cancer progression
Oncogene (2022)
-
Clinical actionability of molecular targets in endometrial cancer
Nature Reviews Cancer (2019)
-
The evolution of endometrial carcinoma classification through application of immunohistochemistry and molecular diagnostics: past, present and future
Virchows Archiv (2018)