Abstract
Pyogenic granuloma, also called lobular capillary hemangioma, is a condition usually occurring in skin or mucosa and often related to prior local trauma or pregnancy. However, the etiopathogenesis of pyogenic granuloma is poorly understood and whether pyogenic granuloma being a reactive process or a tumor is unknown. In an attempt to clarify this issue, we performed genome-wide transcriptional profiling of laser-captured vessels from pyogenic granuloma and from a richly vascularized tissue, placenta, as well as, from proliferative and involutive hemangiomas. Our study identified a gene signature specific to pyogenic granuloma. In the serial analysis of gene expression (SAGE) database, this signature was linked to ‘white blood cells monocytes’. It also demonstrated high enrichment for gene ontology terms corresponding to ‘vasculature development’ and ‘regulation of blood pressure’. This signature included genes of the nitric oxide pathway alongside genes related to hypoxia-induced angiogenesis and vascular injury, three conditions biologically interconnected. Finally, one of the genes specifically associated with pyogenic granuloma was FLT4, a tyrosine-kinase receptor related to pathological angiogenesis. All together, these data advocate for pyogenic granuloma to be a reactive lesion resulting from tissue injury, followed by an impaired wound healing response, during which vascular growth is driven by FLT4 and the nitric oxide pathway.
Similar content being viewed by others
Main
Vascular lesions remain a challenging area for pathologists. It is not uncommon to see many types of vascular lesions reported under the generic term of ‘hemangioma’. It is possibly a residual influence from an early classification published by Virchow1 that categorized vascular lesions according to the type of vessels they are constituted of. Currently, the International Society for the Study of Vascular Anomalies aims at classifying them according to their clinico-pathological characteristics and as such, divides them into vascular malformations and vascular tumors, a suggestion initially made by Mulliken and Glowacki.2 However, this classification does not take into account vascular lesions presumed to be reactive such as reactive angioendotheliomatosis and possibly pyogenic granuloma, also known as lobular capillary hemangioma.
Identification of genetic abnormalities in various vascular malformations has resulted in a great improvement of their classification and in understanding their pathogenesis (for review, see Boon et al3). Comparable advances have not yet been made for pyogenic granuloma. This is attested to by the use of two different terms to define the same lesion: ‘pyogenic granuloma’, which implies a reactive condition, and ‘lobular capillary hemangioma’, which is indicative of a tumor. By comparing genome-wide transcriptional profiling of laser-captured, formalin-fixed and paraffin-embedded micro-dissected vessels of pyogenic granuloma, infantile hemangiomas and placenta, we attempted to better understand the pathogenesis of pyogenic granuloma.
Unsupervised statistical analyses demonstrated vessels of pyogenic granuloma to have a genetic profile distinct from those of infantile hemangiomas and placenta. Supervised analyses isolated a gene signature more specifically related to pyogenic granuloma. By analyzing gene ontology, terms this signature was linked to, serial analysis of gene expression tissues it was associated with, pathways it was affiliated with, and also the biological functions described for the genes constituting the signature. We advocate that pyogenic granuloma results from tissue injury, followed by an impaired wound healing process, during which vascular growth is driven by FLT4 and the nitric oxide pathway.
Materials and methods
Samples
After an initial control step for RNA quantity and quality, capillaries from formalin-fixed and paraffin-embedded normal term placenta (n=3), proliferative infantile hemangioma (n=3), involuting infantile hemangioma (n=4) and pyogenic granuloma (n=4) were laser captured on the Veritas Microdissection Instrument (Applied Biosystems, Carlsbad, CA, USA), as previously described.4
Array Hybridization
RNA was extracted and amplified using the Paradise Reagent System Kit according to the manufacturer’s recommendations (Paradise Reagent System Kit, Applied Biosystems). The amplified and purified cDNAs were then submitted to the Harvard–Partner Center for Genetics and Genomics Facility (www.hpcgg.org). Samples were labeled with biotinylated probes using Bioarray High Yield transcription kit following manufacturer’s protocol (Enzo Biochemical, New York, NY, USA). Their concentration was determined by UV absorbance utilizing a Bio-Tek Plate Reader (Bio-Tek Instruments, Winooski, VT, USA). After fragmentation, 20 μg of each biotinylated cRNA preparation was hybridized to the Affymetrix Human X3P GeneChip Array (http://www.affymetrix.com/products/arrays/specific/x3p.affxArrays) according to the manufacturer’s protocol. Microarrays were washed and stained on a Model 450 Fluidics station controlled by the Affymetrix GeneChip Operating System (GCOS). Images from the scanned chips were processed using an Affymetrix Model 7000 scanner with autoloader. The Affymetrix GCOS v1.3 operating system controls the Model 7000 scanner and data acquisition functions (www.affymetrix.com). Image files were downloaded, imported and analyzed.
Bioinformatics
Only transcripts present were considered for statistical analyses (n=21 318). They were selected by excluding the transcripts that had an intensity value of <19, in less than nine out of the 14 samples studied. Those with a fold difference >2 were further studied. To identify these genes, a comparison was made between the mean of the intensity values from the transcripts of pyogenic granuloma and one of the other tissue/lesions tested. Additionally, the transcripts with a s.d. >1.8 were analysed. To isolate these, the median of the signal intensity of each transcript was defined for each group of lesions or tissue studied. The mean of these medians was then calculated, and subsequently used to divide the signal intensity of the individual transcript. Afterwards a log 2 conversion was performed, and the s.d. of each transcript was defined.
Transcripts with a s.d. >1.8 were submitted for further statistical analyses performed on the TIGR MultiExperiment Viewer platform (MeV4.6, http://www.tm4.org/mev/5). Correspondence analysis was performed. This analysis permits a statistical visualization of the relationship between transcript profiles and samples and was run using the three most informative χ2-values. Analysis of variance was also employed, based on 5000 hazardous permutations. This allowed for the identification of genes more specifically associated to each one of the four tissue/lesions analyzed. To identify genes more specifically related to pyogenic granuloma, studies were made comparing two groups of transcripts: one including transcripts from pyogenic granuloma, and the other, from infantile hemangiomas and placenta combined. Between these two groups, a t-test was run on the MeV platform using 5000 hazardous permutations, a P-value of 0.001 and a Bonferroni adjusted correction. In-house filters were also designed to detect transcripts specifically over- and underexpressed in all pyogenic granuloma samples compared with the other groups.
Finally, the identified transcripts were annotated using the DAVID platform (http://david.abcc.ncifcrf.gov/), the UCSC Genome Bioinformatics database (http://genome.ucsc.edu/), the Standford Microarray database (http://smd.standford.edu/) and PubMed (http://ncbi.nlm.nih.gov/pubmed/).
Immunohistochemistry
Formalin-fixed and paraffin-embedded sections from pyogenic granulomas (n=8), placentas (n=3), proliferative infantile hemangiomas (n=3), involutive infantile hemangiomas (n=6), kaposiform hemangioendotheliomas (n=2), epithelioid hemangioendothelioma (n=1), reactive angioendotheliomatosis (n=1), venous malformation (n=1), spindle cell hemangioma (n=1) and inflammatory granulation tissue (n=4) were stained using the Ventana Discovery XT automated slide processor for an antibody directed against PLVAP (Rabbit Polyclonal, Sigma, dilution: 1/100) and the secondary OmniMap DAB anti-Rabbit detection kits (Ventana Medical Systems, Tucson, AZ, USA). Standard quality control procedures were undertaken to optimize antigen retrieval, primary antibody dilution, secondary antibody detection and other factors for both ‘signal and noise’.
Results
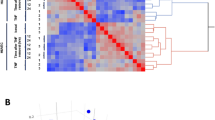
Correspondence analysis was one of the studies performed based on the genome-wide transcriptional profiling of laser capture formalin-fixed and paraffin-embedded micro-dissected vessels of pyogenic granuloma, placenta, proliferative and involutive infantile hemangiomas. This allowed a better understanding of the relationship between the gene expression of the 14 analyzed samples of the four different types of lesions and tissue (Figure 1). This analysis revealed that the genes of similar lesions or tissue clustered together and thus, formed four groups, but with a discrete overlap between proliferative and involutive infantile hemangiomas.
A one-way analysis of variance was then carried out to identify the transcripts implicated in this partition into four groups. It identified 43 transcripts corresponding to 37 known genes. These were used for subsequent unsupervised hierarchical clustering (Figure 2). The derived phylogenetic tree demonstrated that the samples to divide first into two groups: one comprised of the placenta and the other consisting of pyogenic granuloma and infantile hemangiomas. A second division separated this latter group into pyogenic granuloma on one hand and hemangiomas on the other. A final split separated hemangiomas according to their phase, proliferative or involutive, with one proliferative hemangioma mis-clustered into involutive hemangiomas. For the genes identified by the analysis of variance, unsupervised hierarchical clustering assembled them into four groups: A–D (Figure 2). Group A contained eight genes highly upregulated in the placenta. They showed high enrichment scores for the Gene Ontology terms ‘extracellular region’ and ‘female pregnancy’. An association with ‘placenta normal’ in Expressed Sequence Tags database was detected. Group B, comprised 15 genes upregulated in placenta and infantile hemangiomas. They were linked to Gene Ontology terms including ‘plasma membrane’, ‘regulation of system process’ and ‘cell surface receptor-linked signal transduction’. Genes of group C were related to pyogenic granuloma and to ‘vascular, normal liver’ in the Serial Analysis of Gene Expression database. The last 10 genes, forming group D, were common to vessels of proliferative and involutive infantile hemangiomas. They were associated with the Gene Ontology term corresponding to ‘intrinsic to membrane’.
Heat-map of hierarchical clustering performed on genes identified by an analysis of variance. PG: pyogenic granuloma, PL: placenta, IH: involutive hemangioma and PH: proliferative hemangioma. (a) Genes specifically related to placenta. (b) Genes associated with placenta, proliferative and involutive infantile hemangiomas. (c) Genes more specifically linked to pyogenic granuloma. (d) Genes more specifically linked to proliferative and involutive infantile hemangiomas.
To identify genes specifically related to the vessels of pyogenic granuloma, a t-test was performed comparing the genes expressed in pyogenic granuloma to those of the placenta and hemangiomas combined. This analysis yielded a list of 31 differentially expressed genes (Figure 3a). According to the Serial Analysis of Gene Expression database, these genes were related to ‘vascular hemangioma’, ‘placenta’ and ‘vascular normal liver’. In the Kegg pathways, they were associated with ‘neuroactive ligand-receptor interaction’ while in Gene Ontology, they were classified into ‘vascular process in circulatory system’, ‘negative regulation of blood pressure’ and ‘integral to membrane’. Additionally, two of the genes, BDKRB2 and PDE2A, belong to nitric oxide pathway.
(a) Heat-map of significant genes isolated by a t-test comparing genes expressed in pyogenic granuloma to the ones in placenta and infantile hemangiomas combined. (b) Heat-map of significant genes isolated by in-house algorithms comparing genes expressed in pyogenic granuloma to those in placenta and infantile hemangiomas combined. (c) Heat-map of the 10 genes common to the t-test and in-house algorithm as specifically expressed in pyogenic granuloma compared with placenta and infantile hemangiomas combined.
An in-house algorithm was designed in MS Excel with the intent to determine the genes specifically up- and downregulated in all pyogenic granulomas when compared with placenta and infantile hemangiomas combined. It recognized 45 such transcripts, corresponding to 43 genes with nine downregulated and 34 upregulated in pyogenic granuloma (Figure 3b). With the exception of one, ECE1, the genes selected by this method corresponded exactly to the ones isolated by a fold difference greater than two (Figure 4). In the Serial Analysis of Gene Expression database, these genes were associated with ‘vascular endothelium’, ‘white blood cells monocytes’, ‘vascular hemangioma’ and various cancers. They also showed high enrichment for Gene Ontology terms corresponding to ‘embryonic morphogenesis’, ‘blood vessel morphogenesis’, ‘vasculature development’, ‘regulation of blood pressure’, ‘response to hypoxia’ and ‘cell surface receptor-linked signal transduction’. This gene signature was affiliated with five Kegg pathways: focal adhesion with FLT-1, FLT4 and PARVB; purine metabolism with ENTPD1 and PDE2A; endocytosis with EHD4 and FLT-1; chemokine signaling pathway with GNG7 and PREX1; and cytokine–cytokine receptor interaction with FLT-1 and FLT4. In Biocarta, two genes were linked to ‘actions of nitric oxide in the heart’: BDKRB2 and PDE2A. Finally, among these 43 genes, 10 were also identified by the t-test with six being upregulated in pyogenic granuloma (FLT4, ST3GAL6, PDE2A, BDKRB2, PLVAP and SPTBN5) and four downregulated (H19, CGNL1, FOXF1 and MEG3) (Figure 3c). Their fold differences ranged from 3.2 to 12.7 and from 4.2 to 19.8, respectively (Figure 4).
Immunohistochemistry
All pyogenic granulomas showed moderate to strong staining for PLVAP in the endothelium of all capillaries (n=8, Figure 5a). Placentas show mild-to-moderate endothelial staining in a few capillaries in two cases and most capillaries in one case (n=3, Figure 5b). Proliferative infantile hemangiomas had very weak staining in 10–50% of capillaries (n=3, Figure 5c). Involutive infantile hemangiomas immuno-positivity ranged from negative to moderately positive from a few capillaries to as many as 30% (n=6, Figure 5d). Kaposiform hemangioendotheliomas were moderately to strongly positive in all capillaries (n=2). Epithelioid hemangioendothelioma was negative (n=1). Reactive angioendotheliomatosis was moderately positive (n=1). Venous malformation was focally weakly positive (n=1). Spindle cell hemangioma was moderately focally positive (n=1). Inflammatory granulation tissue showed moderate-to-strong staining in all capillaries (n=4). In all of the above, a variable number of arteries and veins stained weakly to strongly, the veins generally more than the arteries, as well as, in a survey of normal organs from an autopsy. In addition, most if not all capillaries stained strongly.
Discussion
Pyogenic granuloma, also called lobular capillary hemangioma, is a vascular proliferative lesion most commonly affecting skin or mucosa of children and young adults.6 On rare occasions, it has been reported in organs such as liver7 or central nervous system.8 They have also been observed in the lumen of vessels9 or in vascular lesions such as arteriovenous malformation10 and capillary malformation.11 Their pathogenesis is still poorly understood. Links between pyogenic granuloma development and pregnancy or trauma have been reported, and suggested as causative factor.12, 13 To improve our understanding of these lesions, we analyzed genome-wide transcriptional profiles of laser-captured micro-dissected vessels from pyogenic granuloma and from three richly vascularized entities, namely placenta, proliferative, and involutive infantile hemangiomas.
In the first part of our study, we attempted to better understand the relationship between the four tested entities. The different statistical analyses performed identified sets of genes specific for vessels of each tissue type, alongside sets of genes common to vessels of different tissue types. This could be expected considering the design of the study that might schematically be phrased as: ‘studying one tissue type, vessel, in various conditions’. Vessels of placenta and of infantile hemangiomas nicely illustrated this genetic similarity/dissimilarity. Indeed, correspondence analysis revealed vessels of the placenta to have a genetic signature distinct from that of all the other tested tissues. This finding was reinforced by the analysis of variance that identified a group of eight genes only upregulated in the placenta. Simultaneously, the latter test detected an additional set of 15 genes shared by the placenta and infantile hemangiomas, a likely finding as genetic similarities between infantile hemangioma and placenta are known.14, 15
Among these 15 genes, one observed genes implicated in angiogenesis, CLU16 and FST17 as well as imprinted genes, such as DIO3 and DLK1. The latter are part of the mir-379/mir-656 miRNA cluster, a chromosomal region that appears increasingly more relevant to normal placental development17, 18, 19 and fetal growth,20 stem cell differentiation21 and various pathological conditions.22, 23, 24 Further statistical analyses identified additional imprinted genes as specifically related to the placenta and hemangioma: PLAGL1, DIO3OS, TFPI2, GRB14, H19 and MEG3. If the role of imprinted genes in placental development was already well established, it is, to our knowledge, the first assessment of them in infantile hemangiomas.
Between proliferative and involutive infantile hemangiomas, genetic similarity was recognized, with the identification by the analysis of variance of a cluster of 10 genes related to both of them. These genes were linked to the Gene Ontology term corresponding to ‘intrinsic to membrane’ and the two genes, ADCYAP1R1 and PTGER3, are known to have a role in angiogenesis.25, 26 Finding some degree of genetic similarity between proliferative and involutive infantile hemangiomas was not surprising as they are only two different time points in the natural evolution of these lesions. Our statistical analyses were not designed to unravel the differences between the stages of infantile hemangiomas but, studies that did, determined them to differ with regard to the genes of the chronic inflammatory response and of the NOTCH pathway.4, 27
For pyogenic granuloma, correspondence analysis and analysis of variance revealed only dissimilarities with the other tissues tested. Such specific genetic behavior of vessels from pyogenic granuloma was not surprising, although not previously described. Indeed, pyogenic granuloma is clinically dissimilar from placenta and from infantile hemangiomas. For example, pyogenic granuloma almost never regresses spontaneously, in contrast to infantile hemangiomas.6
Because of these results, and to better understand the pathogenesis of pyogenic granuloma, we compared their profiles to those of placenta and hemangiomas combined, using the t-test, in-house algorithms and fold differences. It allowed identification of genes specifically related to pyogenic granuloma. FLT-1 and FLT4, two tyrosine-kinase receptors, related to angiogenesis and vasculogenesis and to lymphangiogenesis, respectively. Interestingly, FLT4 has been recently implicated in angiogenesis of tumors and wounds28 (for review see Bahram and Claesson-Welsh29). In this situation, and in contrast to FLT-1, signaling mediated through FLT4 does not require activation by any of its recognized ligands, VEGFB or VEGFC.30 Finally, FLT4 missense somatic mutations have been found in infantile hemangiomas,31 further implicating this gene in the pathogenesis of pyogenic granuloma.
ENG is specifically upregulated in pyogenic granuloma. Mutations in it cause hereditary hemorrhagic telangiectasia type 1, also known as Osler–Rendu–Weber disease, an autosomal dominant multisystemic vascular dysplasia (MIM: 187300, 108010). This gene codes for a major glycoprotein of the vascular endothelium implicated in both vasculogenesis and angiogenesis.
Other genes linked to pyogenic granuloma, EMCN, PODXL and ENTPD1, are related to hematopoietic stem cells,32, 33, 34 and such would favor pyogenic granuloma to be linked to vasculogenesis. But, as discussed for infantile hemangiomas, pyogenic granuloma could similarly be viewed as the result of a vascular program concurrently associating with vasculogenesis and angiogenesis.35 ENTPD1 has been linked to hypoxia-induced angiogenesis as BDKRB2 and WARS, two other genes upregulated in pyogenic granuloma.36, 37, 38 The angiogenic action of BDKRB2 appears to result from primary intracellular production of nitric oxide.37 PDE2A is another gene upregulated in pyogenic granuloma and implicated in the nitric oxide pathway39 (Biocarta). Interestingly, the role of nitric oxide in the post-ischemic revascularization process has been well-documented.40, 41, 42 Such findings highlight the role of the nitric oxide pathway in pyogenic granuloma development, possibly secondary to a hypoxic event.
In addition, the nitric oxide pathway has also been related to vascular injury.43, 44 Many genes specific to pyogenic granuloma are, at least by their functions, related to vascular injury, wound healing process or cancer angiogenesis: BDKRB2,45, 46, 47 NTPDase1,48 ENG,49, 50 FLT4,28 ST3GAL6,51 NDST1,52 HSPG253 and HOXD3.54 Moreover, all these genes have a fold difference >2 in pyogenic granuloma when compared with other tissue tested, further reinforcing the connection between pyogenic granuloma and vascular injury.
Immunohistochemistry directed against PLVAP was used to validate the array data. PLVAP was chosen because this gene was found to be specifically upregulated in pyogenic granuloma by all our statistical analyses, and having the second highest fold difference. Recently, the link between PLVAP and an antibody used for years as a vascular maker, the ‘Pathologische Anatomie Leiden-endothelium’ antibody, was described.55 It was therefore not surprising to observe immuno-positivity of endothelial cells in various tissues, but the staining intensity and its frequency were highest in pyogenic granuloma, thus confirming the array data.
Finally, inflammatory granulation tissue, a known reactive lesion, was also tested for PLVAP reactivity. It demonstrated PLVAP positivity, but to a lesser extent in pyogenic granulomas. Interestingly, PLVAP has recently been demonstrated to play a role in leukocyte transendothelial migration.55 All this reinforces the link between pyogenic granuloma and tissue insult.
In conclusion, genome-wide profiling analyses of micro-dissected vessels from formalin-fixed and paraffin-embedded placenta, infantile hemangiomas and pyogenic granuloma demonstrated genetic similarity/dissimilarity between the placenta, proliferative and involutive infantile hemangiomas, as well as dissimilarity with the vessels of pyogenic granuloma. Ascertainment of the genes specifically associated with pyogenic granuloma leads us to believe that it results from an injury followed by an impaired wound healing process associated with vascular growth driven by FLT4 and the nitric oxide pathway. Thus, this favors pyogenic granuloma to be a reactive lesion and not a tumor.
References
Virchow R . Angiome In: Die krankhaften Geschwülste. August Hirschwald: Berlin, 1863, vol 3, pp306.
Mulliken JB, Glowacki J . Classification of pediatric vascular lesions. Plast Reconstr Surg 1982;70:120–121.
Boon LM, Ballieux F, Vikkula M . Pathogenesis of vascular anomalies. Clin Plast Surg 2011;38:7–19.
Calicchio ML, Collins T, Kozakewich HP . Identification of signaling systems in proliferating and involuting phase infantile hemangiomas by genome-wide transcriptional profiling. Am J Pathol 2009;174:1638–1649.
Saeed AI, Sharov V, White J et al TM4: a free, open-source system for microarray data management and analysis. Biotechniques 2003;34:374–378.
Patrice SJ, Wiss K, Mulliken JB . Pyogenic granuloma (lobular capillary hemangioma): a clinicopathologic study of 178 cases. Pediatr Dermatol 1991;8:267–276.
Abaalkhail F, Castonguay M, Driman DK et al Lobular capillary hemangioma of the liver. Hepatobiliary Pancreat Dis Int 2009;8:323–325.
Abe M, Misago N, Tanaka S et al Capillary hemangioma of the central nervous system: a comparative study with lobular capillary hemangioma of the skin. Acta Neuropathol 2005;109:151–158.
Cooper PH, McAllister HA, Helwig EB . Intravenous pyogenic granuloma. A study of 18 cases. Am J Surg Pathol 1979;3:221–228.
Hung CH, Kuo HW, Chiu YK et al Intravascular pyogenic granuloma arising in an acquired arteriovenous malformation: report of a case and review of the literature. Dermatol Surg 2004;30:1050–1053.
Chen D, Hu XJ, Lin XX et al Nodules arising within port-wine stains: a clinicopathologic study of 31 cases. Am J Dermatopathol 2011;33:144–151.
Henry F, Quatresooz P, Valverde-Lopez JC et al Blood vessel changes during pregnancy: a review. Am J Clin Dermatol 2006;7:65–69.
Lin RL, Janniger CK . Pyogenic granuloma. Cutis 2004;74:229–233.
North PE, Waner M, Mizeracki A et al A unique microvascular phenotype shared by juvenile hemangiomas and human placenta. Arch Dermatol 2001;137:559–570.
Barnes CM, Huang S, Kaipainen A et al Evidence by molecular profiling for a placental origin of infantile hemangioma. Proc Natl Acad Sci USA 2005;102:19097–19102.
Jackson JK, Gleave ME, Gleave J et al The inhibition of angiogenesis by antisense oligonucleotides to clusterin. Angiogenesis 2005;8:229–238.
Lin SY, Craythorn RG, O’Connor AE et al Female infertility and disrupted angiogenesis are actions of specific follistatin isoforms. Mol Endocrinol 2008;22:415–429.
Fowden AL, Sibley C, Reik W et al Imprinted genes, placental development and fetal growth. Horm Res 2006;65 (Suppl 3):50–58.
Glazov EA, McWilliam S, Barris WC et al Origin, evolution, and biological role of miRNA cluster in DLK-DIO3 genomic region in placental mammals. Mol Biol Evol 2008;25:939–948.
Frost JM, Moore GE . The importance of imprinting in the human placenta. PLoS Genet 2010;6:e1001015.
Liu L, Luo GZ, Yang W et al Activation of the imprinted Dlk1-Dio3 region correlates with pluripotency levels of mouse stem cells. J Biol Chem 2010;285:19483–19490.
Gardiner E, Beveridge NJ, Wu JQ et al Imprinted DLK1-DIO3 region of 14q32 defines a schizophrenia-associated miRNA signature in peripheral blood mononuclear cells. Mol Psychiatry 2011;17:827–840.
Luk JM, Burchard J, Zhang C et al DLK1-DIO3 genomic imprinted microRNA cluster at 14q32.2 defines a stemlike subtype of hepatocellular carcinoma associated with poor survival. J Biol Chem 2011;286:30706–30713.
Kagami M, Sekita Y, Nishimura G et al Deletions and epimutations affecting the human 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat Genet 2008;40:237–242.
Castorina A, Giunta S, Mazzone V et al Effects of PACAP and VIP on hyperglycemia-induced proliferation in murine microvascular endothelial cells. Peptides 2010;31:2276–2283.
Amano H, Hayashi I, Endo H et al Host prostaglandin E(2)-EP3 signaling regulates tumor-associated angiogenesis and tumor growth. J Exp Med 2003;197:221–232.
Wu JK, Adepoju O, De Silva D et al A switch in Notch gene expression parallels stem cell to endothelial transition in infantile hemangioma. Angiogenesis 2010;13:15–23.
Tammela T, Zarkada G, Wallgard E et al Blocking VEGFR-3 suppresses angiogenic sprouting and vascular network formation. Nature 2008;454:656–660.
Bahram F, Claesson-Welsh L . VEGF-mediated signal transduction in lymphatic endothelial cells. Pathophysiology 2010;17:253–261.
Zhang L, Zhou F, Han W et al VEGFR-3 ligand-binding and kinase activity are required for lymphangiogenesis but not for angiogenesis. Cell Res 2010;20:1319–1331.
Walter JW, North PE, Waner M et al Somatic mutation of vascular endothelial growth factor receptors in juvenile hemangioma. Genes Chromosomes Cancer 2002;33:295–303.
Matsubara A, Iwama A, Yamazaki S et al Endomucin, a CD34-like sialomucin, marks hematopoietic stem cells throughout development. J Exp Med 2005;202:1483–1492.
Hara T, Nakano Y, Tanaka M et al Identification of podocalyxin-like protein 1 as a novel cell surface marker for hemangioblasts in the murine aorta-gonad-mesonephros region. Immunity 1999;11:567–578.
Ciriza J, Garcia-Ojeda ME . Expression of migration-related genes is progressively upregulated in murine Lineage-Sca-1+c-Kit+ population from the fetal to adult stages of development. Stem Cell Res Ther 2010;1:14.
Boscolo E, Bischoff J . Vasculogenesis in infantile hemangioma. Angiogenesis 2009;12:197–207.
Candinas D, Koyamada N, Miyatake T et al Loss of rat glomerular ATP diphosphohydrolase activity during reperfusion injury is associated with oxidative stress reactions. Thromb Haemost 1996;76:807–812.
Liesmaa I, Leskinen HK, Kokkonen JO et al Hypoxia-induced expression of bradykinin type-2 receptors in endothelial cells triggers NO production, cell migration, and angiogenesis. J Cell Physiol 2009;221:359–366.
Zeng R, Chen YC, Zeng Z et al Effect of mini-tyrosyl-tRNA synthetase/mini-tryptophanyl-tRNA synthetase on ischemic angiogenesis in rats: proliferation and migration of endothelial cells. Heart Vessels 2011;26:69–80.
Venkatesha S, Toporsian M, Lam C et al Soluble endoglin contributes to the pathogenesis of preeclampsia. Nat Med 2006;12:642–649.
Luque Contreras D, Vargas Robles H, Romo E et al The role of nitric oxide in the post-ischemic revascularization process. Pharmacol Ther 2006;112:553–563.
Mendoza MG, Robles HV, Romo E et al Nitric oxide-dependent neovascularization role in the lower extremity disease. Curr Pharm Des 2007;13:3591–3596.
Alef MJ, Vallabhaneni R, Carchman E et al Nitrite-generated NO circumvents dysregulated arginine/NOS signaling to protect against intimal hyperplasia in Sprague-Dawley rats. J Clin Invest 2011;121:1646–1656.
Kibbe M, Billiar T, Tzeng E . Inducible nitric oxide synthase and vascular injury. Cardiovasc Res 1999;43:650–657.
Thom SR, Fisher D, Xu YA et al Role of nitric oxide-derived oxidants in vascular injury from carbon monoxide in the rat. Am J Physiol 1999;276:H984–H992.
Chachisvilis M, Zhang YL, Frangos JA . G protein-coupled receptors sense fluid shear stress in endothelial cells. Proc Natl Acad Sci USA 2006;103:15463–15468.
Plendl J, Snyman C, Naidoo S et al Expression of tissue kallikrein and kinin receptors in angiogenic microvascular endothelial cells. Biol Chem 2000;381:1103–1115.
Ikeda Y, Hayashi I, Kamoshita E et al Host stromal bradykinin B2 receptor signaling facilitates tumor-associated angiogenesis and tumor growth. Cancer Res 2004;64:5178–5185.
Behdad A, Sun X, Khalpey Z et al Vascular smooth muscle cell expression of ectonucleotidase CD39 (ENTPD1) is required for neointimal formation in mice. Purinergic Signal 2009;5:335–342.
van Laake LW, van den Driesche S, Post S et al Endoglin has a crucial role in blood cell-mediated vascular repair. Circulation 2006;114:2288–2297.
Torsney E, Charlton R, Parums D et al Inducible expression of human endoglin during inflammation and wound healing in vivo. Inflamm Res 2002;51:464–470.
Gracheva EV, Samovilova NN, Golovanova NK et al Sialyltransferase activity of human plasma and aortic intima is enhanced in atherosclerosis. Biochem Biophys Acta 2002;1586:123–128.
Adhikari N, Rusch M, Mariash A et al Alterations in heparan sulfate in the vessel in response to vascular injury in the mouse. J Cardiovasc Transl Res 2008;1:236–240.
Zhou Z, Wang J, Cao R et al Impaired angiogenesis, delayed wound healing and retarded tumor growth in perlecan heparan sulfate-deficient mice. Cancer Res 2004;64:4699–4702.
Hansen SL, Myers CA, Charboneau A et al HoxD3 accelerates wound healing in diabetic mice. Am J Pathol 2003;163:2421–2431.
Keuschnigg J, Henttinen T, Auvinen K et al The prototype endothelial marker PAL-E is a leukocyte trafficking molecule. Blood 2009;114:478–484.
Acknowledgements
These studies were supported in part by NIH RO1 HL35716 and funds from the Wolbach Chair at Harvard Medical School. We would like to thank Tucker Collins, MD, PhD (1952-2007), our mentor and friend, for his support and enthusiasm.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Godfraind, C., Calicchio, M. & Kozakewich, H. Pyogenic granuloma, an impaired wound healing process, linked to vascular growth driven by FLT4 and the nitric oxide pathway. Mod Pathol 26, 247–255 (2013). https://doi.org/10.1038/modpathol.2012.148
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2012.148
Keywords
This article is cited by
-
Efficacy of Nd-YAG laser for treatment of pyogenic granuloma on the fingers and toes
Lasers in Medical Science (2019)