Abstract
Immunohistochemistry is increasingly utilized to differentiate lung adenocarcinoma and squamous cell carcinoma. However, detailed analysis of coexpression profiles of commonly used markers in large series of whole-tissue sections is lacking. Furthermore, the optimal diagnostic algorithm, particularly the minimal-marker combination, is not firmly established. We therefore studied whole-tissue sections of resected adenocarcinoma and squamous cell carcinoma (n=315) with markers commonly used to identify adenocarcinoma (TTF-1) and squamous cell carcinoma (p63, CK5/6, 34βE12), and prospectively validated the devised algorithm in morphologically unclassifiable small biopsy/cytology specimens (n=38). Analysis of whole-tissue sections showed that squamous cell carcinoma had a highly consistent immunoprofile (TTF-1-negative and p63/CK5/6/34βE12-diffuse) with only rare variation. In contrast, adenocarcinoma showed significant immunoheterogenetity for all ‘squamous markers’ (p63 (32%), CK5/6 (18%), 34βE12 (82%)) and TTF-1 (89%). As a single marker, only diffuse TTF-1 was specific for adenocarcinoma whereas none of the ‘squamous markers,’ even if diffuse, were entirely specific for squamous cell carcinoma. In contrast, coexpression profiles of TTF-1/p63 had only minimal overlap between adenocarcinoma and squamous cell carcinoma, and there was no overlap if CK5/6 was added as a third marker. An algorithm was devised in which TTF-1/p63 were used as the first-line panel, and CK5/6 was added for rare indeterminate cases. Prospective validation of this algorithm in small specimens showed 100% accuracy of adenocarcinoma vs squamous cell carcinoma prediction as determined by subsequent resection. In conclusion, although reactivity for ‘squamous markers’ is common in lung adenocarcinoma, a two-marker panel of TTF-1/p63 is sufficient for subtyping of the majority of tumors as adenocarcinomas vs squamous cell carcinoma, and addition of CK5/6 is needed in only a small subset of cases. This simple algorithm achieves excellent accuracy in small specimens while conserving the tissue for potential predictive marker testing, which is now an essential consideration in advanced lung cancer specimens.
Similar content being viewed by others
Main
Adenocarcinoma and squamous cell carcinoma are the two major subtypes of non-small cell lung carcinoma. Until recently, therapeutic approaches to non-small cell lung carcinoma were largely guided by tumor stage, and there was no difference in treatment for adenocarcinoma vs squamous cell carcinoma. This monolithic approach to non-small cell lung carcinoma has dramatically changed in the last few years as a result of three major advances in thoracic medical oncology for advanced disease. These include (1) EGFR-targeted therapies, erlotinib (Tarceva) and gefitinib (Iressa), which are currently recommended as the first-line treatment for non-small cell lung carcinoma with EGFR mutations, and these mutations occur primarily in adenocarcinoma,1 (2) bevacizumab (Avastin), which is contraindicated in squamous cell carcinoma due to the risk of pulmonary hemorrhage,2 and (3) pemetrexed (Alimta), which is also contraindicated in squamous cell carcinoma due to the lack of effectiveness.3 In addition, recently discovered EML4-ALK translocation, which predicts sensitivity to a targeted agent PF-02341066 (Crizotinib), also occurs specifically in adenocarcinoma.4 Other important molecular differences between adenocarcinoma and squamous cell carcinoma are increasingly identified,5, 6 suggesting that future targeted therapies will be increasingly ‘histology specific.’ Selection of patients for appropriate molecular tests and histology-based therapies necessitates accurate pathologic distinction of adenocarcinoma vs squamous cell carcinoma.
In most cases, the distinction of adenocarcinoma and squamous cell carcinoma is readily achieved based on standard morphologic criteria,7 with keratinization and intercellular bridges representing hallmarks of squamous cell carcinoma and glandular architecture (in the form of acini, papillae, micropapillae, or cytoplasmic mucin) representing the hallmarks of adenocarcinoma. However, distinction can be difficult in some poorly differentiated tumors, where defining glandular or squamous features are subtle or focal. This issue is particularly amplified in small specimens (small biopsies and cytology) where focal evidence of morphologic differentiation may not be represented as a result of scant cellularity, crush artifact, or cell dispersal. Because ∼70% of non-small cell lung carcinoma present at an unresectable stage, the only diagnostic material guiding systemic therapy in the majority of such patients are small specimens. Until recently, a non-committal diagnosis of non-small cell lung carcinoma—not otherwise specified was widely advocated as a general approach to small specimens because of the inability of morphology to distinguish some poorly differentiated tumors combined with the lack of clinical significance.8 This practice trend is reflected in a 20–40% rate of non-small cell lung carcinoma—not otherwise specified diagnoses in small specimens in recent years, and a near doubling in the use of this category between 1989 and 2006.9 Recently, as a result of the above so-called ‘histology-based’ molecular and therapeutic advances, there has been a major paradigm shift in the approach to pathologic diagnosis of non-small cell carcinoma with a new emphasis now placed on specific and accurate non-small cell carcinoma subtyping in small specimens.10, 11 This new approach to non-small cell carcinoma diagnosis is advocated in the recent IASLC/ATS/ERS lung adenocarcinoma classification.12
In parallel with the above clinical and molecular progress, the major advance in thoracic pathology in recent years has been the growing evidence that immunohistochemistry is a highly effective ancillary tool for distinguishing adenocarcinoma and squamous cell carcinoma. Although the optimal diagnostic algorithm is not firmly established, recent studies show that immunohistochemistry increases accuracy and reproducibility, and minimizes the rate of non-small cell carcinoma—not otherwise specified diagnoses in small specimens.13, 14, 15, 16, 17 We recently showed that the rate of adenocarcinoma and squamous cell carcinoma unclassified by preoperative cytology in clinical practice with routine utilization of immunohistochemistry is low (3%).18
Common markers used for non-small cell carcinoma subtyping include TTF-1 for adenocarcinoma vs p63 and high-molecular weight keratins (CK5/6 and 34βE12/CK903) for squamous cell carcinoma. Other markers are further addressed in the Discussion. The difficulty with all of the markers has been that none are individually entirely tumor-type sensitive and specific. Furthermore, highly variable results have been reported over the years, particularly for the prevalence of ‘squamous markers’ in adenocarcinoma (eg the reported prevalence of p63 in adenocarcinoma has ranged from 0 to 65%19, 20, 21, 22).
Given the recent insights into the clinical and biological significance of non-small cell carcinoma subtypes, several recent studies have analyzed the utility of various combinations of markers for the distinction of adenocarcinoma and squamous cell carcinoma. These studies were performed in small biopsies,14, 15, 16 cytology,17, 23 and tissue microarrays,24, 25, 26 and the proposed algorithms include between four- and six-marker panels. Although most studies agree on the inclusion of TTF-1 and p63 in the panel, there is no agreement on the role of additional markers. The consideration of a minimal panel is critical for specimens with low cellularity where it may be feasible to perform only a limited number of immunostains and because of the growing need to conserve scant starting material for predictive marker testing.27 The other uncertainty in several recent studies is with interpretation of unusual coexpression profiles (such as TTF-1/p63 double positive). Therefore, the other goal of the study was to review in detail coexpression profiles of commonly used markers in whole-tissue sections. In contrast to algorithms derived from limited tissue samples (biopsies, cytology, tissue microarrays), where the distribution of markers with focal reactivity may be underestimated, analysis of whole-tissue sections reflects the widest spectrum of markers reactivities and possible coexpression patterns. We therefore undertook the approach of analyzing a limited panel of four commonly used markers, which are widely available in pathology laboratories (TTF-1, p63, CK5/6, 34βE12) in a large series of whole-tissue sections (n=315) to devise a minimal-marker algorithm, which we also prospectively validated in a series of small biopsy/cytology specimens (n=38).
Materials and methods
Study Design and Specimen Characteristics
Whole-tissue sections from 315 consecutively resected adenocarcinoma (n=200) and squamous cell carcinoma (n=115) enriched for poorly and moderately differentiated tumors were studied. H&E slides were reviewed by two thoracic pathologists (NR, ALM), and cases with unusual immunoprofiles were additionally reviewed by a third thoracic pathologist (WDT). Tumors were classified and graded by the standard criteria.7 All tumors had at least focal area of definite line of morphologic differentiation. Patients with adenocarcinoma had male-to-female ratio of 0.6, mean age 69.5 years, age range 35–88 years. Surgical procedures included lobectomy (n=131), pneumonectomy (n=1), and wedge resection or segmentectomy (n=68). The distribution of adenocarcinoma grades was well differentiated (n=19, 9%), moderately differentiated (n=104, 52%), and poorly differentiated (n=77, 39%). Patient with squamous cell carcinoma had male-to-female ratio of 1.7, mean age 66.1 years, age range 35–88 years. Surgical procedures included lobectomy (n=62), pneumonectomy (n=12), and wedge resection or segmentectomy (n=41). For squamous cell carcinoma the distribution of tumor grades was well differentiated (n=3, 3%), moderately differentiated (n=53, 46%), and poorly differentiated (n=59, 51%).
For validation, we reviewed the data for 38 consecutive preoperative small biopsies (n=9) and cytology specimens (n=29), which required immunostains to determine non-small cell carcinoma subtype as part of clinical work-up. Included were transthoracic core biopsies (n=7), bronchoscopic biopsies (n=2), transthoracic fine needle aspirates (FNAs) (n=28), and a transbronchial FNA (n=1). For cytology specimens, immunostains were performed on paraffin-embedded cell blocks by the same protocol as surgical specimens. Because the aim was to evaluate the performance of the algorithm in distinguishing adenocarcinoma from squamous cell carcinoma, only cases with a subsequent resection diagnosis of adenocarcinoma or squamous cell carcinoma in at least a component were included.
This study was approved by the Institutional Review Board of Memorial Sloan-Kettering Cancer Center, New York, NY.
Immunohistochemistry
Immunohistochemistry was performed by a standard protocol on Ventana Discovery XT automated stainer (Ventana Medical Systems, Tucson, AZ, USA). Antigen retrieval was performed with CC1 buffer (Cell Conditioning 1; citrate buffer pH 6.0, Ventana). Primary antibodies included TTF-1 (NKX2-1) (SPT24, NovoCastra, 1:50 dilution), p63 (TP63) (4A4, Dako, 1:700 dilution), CK5/6 (D5/16B4, Dako, 1:200 dilution), and 34βE12 (CK903 or K903, Dako, 1:400 dilution). Immunohistochemistry for TTF-1 and p63 was performed on all samples, and CK5/6 and 34βE12 on a subset of cases. In resections, immunoreactivity was scored semiquantitatively by recording percentage of reactive tumor cells. Diffuse reactivity was defined as labeling of ≥50% tumor cells, and focal reactivity as 1–49%. In addition, intensity of reactivity was recorded as weak (1+, less than normal cells); moderate (2+, same as normal cells); and strong (3+, stronger than normal cells). Pneumocytes served as internal controls for TTF-1 reactivity, and bronchial basal cells for p63, CK5/6, and 34βE12 reactivity. H (‘histologic’) scores were derived by multiplying percentage of immunoreactive cells (0 to 100) by intensity score (0, 1+, 2+, 3+), yielding a number between 0 and 300. p63 scoring in squamous cell carcinoma excluded overtly keratinized cells. Small specimens were scored as negative (no reactivity) vs focal (labeling in the minority of cells) vs diffuse (labeling in the majority of cells) without exact quantitation.
Data Analysis
For each marker and combination of markers, sensitivity and specificity were calculated against the gold standard represented by the morphologic diagnosis. Performance of the markers on a continuous scale was analyzed by receiver operator characteristic curves (ROC), in which area under the curve ranges between 0.5 (indicating no predictive value) and 1 (indicating perfect predictive accuracy). The Heatmap was generated using R open-source software (http://www.r-project.org/). Spearman rank correlation (r) was used to assess whether expression of two markers was independent or coordinated, with r value closest to 1.0 indicating a perfect direct relationship, r value close to 0 indicating the absence of a relationship, and r value close to –1.0 indicating a perfect inverse relationship. Significance of associations was analyzed by two-tailed Fisher's exact test for categorical variables or Mann–Whitney t-test for continuous variables. P values of ≤0.05 were considered statistically significant. Percentages were rounded to an integer and may not add up to 100.
Results
One-Marker Expression Profiles in Resected Adenocarcinoma vs Squamous Cell Carcinoma
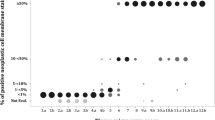
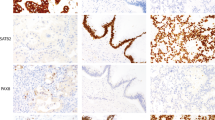
As shown in Table 1, squamous cell carcinoma displayed a highly consistent immunoprofile (TTF-1-negative, p63-diffuse, CK5/6-diffuse, 34βE12-diffuse). In particular, all squamous cell carcinomas were consistently immunoreactive for p63 and 34βE12, and reactivity for these markers in the vast majority of cases was homogeneously strong and diffuse (Figures 1 and 2). In fact, only one case (1%) showed focal rather than diffuse p63 (30% immunoreactive cells), and even reactivity in <75% of tumor cells was highly unusual for p63 occurring in only five cases (4%). Furthermore, the mean percentage of p63-reactive tumor cells did not differ in poorly differentiated vs well–moderately differentiated squamous cell carcinoma (97 vs 94%, P=0.67), supporting that p63 is a highly stable marker in this tumor. The most variable marker in squamous cell carcinoma was CK5/6 in that it could be focal (10%), completely absent (2%), or weak (1+) (13%) (Figures 1 and 2). Furthermore, the mean reactivity for CK5/6 decreased significantly in poorly differentiated compared with well–moderately differentiated squamous cell carcinoma (91 vs 64%, P<0.0001), and 20% of poorly differentiated tumors had ≤10% CK5/6 reactivity. The other deviation from the typical squamous profile was focal reactivity for TTF-1 (SPT24 clone) in four cases (3%) of otherwise classic squamous cell carcinoma both morphologically and based on strong/diffuse expression of all squamous markers.
Comparison of the extent of TTF-1, p63, CK5/6, and 34βE12 immunoreactivity in adenocarcinoma vs squamous cell carcinoma. Scatter dot plots for percentage of immunoreactive cells (a) and H scores (b). Each dot represents reactivity in a single case. Lines and error bars indicate the mean and s.d., respectively. Cases with no reactivity were excluded from this analysis. ADC, adenocarcinoma; SQCC, squamous cell carcinoma.
In contrast to highly homogeneous immunoprofile of squamous cell carcinoma, adenocarcinoma showed significant immunoheterogeneity for all tested markers, including ‘squamous markers’ (p63 (32%), CK5/6 (18%), 34βE12 (82%)), and TTF-1 (89%) (Table 1). Whereas p63 and CK5/6 were only rarely diffuse in adenocarcinoma (5% and 3%, respectively), diffuse reactivity for 34βE12 was extremely common (45%). Similarly, adenocarcinoma only rarely showed intense (3+) reactivity for p63 and CK5/6 (8% and 1% of cases, respectively) (Figure 1), and the combination of diffuse and strong reactivity at the level typical for squamous cell carcinoma (as reflected by H scores) was exceptional for p63 and CK5/6, but common for 34βE12 (Figure 2). TTF-1 reactivity in adenocarcinoma was predominantly bimodal (as previously reported28): either diffusely positive (84%) or completely negative (11%), with focal reactivity seen in only a minority of tumors (6%) (Table 1; Figure 2). TTF-1 distribution in poorly differentiated adenocarcinoma was 78% diffuse, 10% focal, and 12% negative.
Analysis of one-marker sensitivity/specificity (Table 2) showed that none of the four markers was 100% specific for adenocarcinoma vs squamous cell carcinoma if unquantified reactivities were considered (ie any reactivity considered positive). In contrast, diffuse TTF-1 was 100% specific for adenocarcinoma. Similarly, H scores for TTF-1 showed no overlap between adenocarcinoma and squamous cell carcinoma. ROC curve analysis indicated that among ‘squamous markers’ p63 had the best predictive accuracy (as reflected by the highest area under the curve value), CK5/6 was the second best, and 34βE12 was the worst.
Coexpression Profiles in Resected Adenocarcinoma vs Squamous Cell Carcinoma
Because reactivity for individual ‘squamous markers’ did not discriminate between adenocarcinoma and squamous cell carcinoma, we next examined whether there was an overlap in expression of various combinations of markers, particularly the combinations of ‘squamous markers’ with TTF-1. Table 3 shows two-marker coexpression profiles for TTF-1/p63, TTF-1/CK5/6, and TTF-1/34βE12 in adenocarcinoma vs squamous cell carcinoma. The least overlap was seen with TTF-1/p63 panel (discussed below). In contrast, the overlap was substantial for a combination of TTF-1/CK5/6: TTF-1-negative/CK5/6-focal or negative profiles could be seen in either adenocarcinoma (10%) or squamous cell carcinoma (12%). The two-marker overlap was especially high for TTF-1/34βE12 combination in that the majority of TTF-1-negative adenocarcinoma had diffuse 34βE12 (same profile as squamous cell carcinoma), indicating that this profile cannot be used to support squamous cell carcinoma.
TTF-1/p63 coexpression profiles had the following distribution: diffuse TTF-1 (with any level of p63) was seen in 84% of adenocarcinoma vs none of squamous cell carcinoma, whereas TTF-1-negative/p63-diffuse profile was seen in 96% of squamous cell carcinoma vs none of adenocarcinoma. Therefore, these two non-overlapping profiles encompassed the vast majority of adenocarcinoma and squamous cell carcinoma. TTF-1/p63 double-negative profile was seen only in adenocarcinoma (10%) and none of squamous cell carcinoma. Similarly, TTF-1/p63 double-positive tumors were all adenocarcinoma with exception of rare squamous cell carcinoma with focal TTF-1 labeling, but quantitatively TTF-1/p63 ratio in those tumors was low—a ratio not seen in adenocarcinoma. The only overlapping profile was that of negative TTF-1 and focal p63 (‘indeterminate’ profile). This profile was seen only rarely (1% of adenocarcinoma and 1% of squamous cell carcinoma). Addition of CK5/6 (but not 34βE12) correctly stratified these indeterminate cases into squamous cell carcinoma (CK5/6-positive) vs adenocarcinoma (CK5/6-negative). Of note, the extent of p63 in TTF-1-negative adenocarcinoma was minimal (≤10% reactive cells) vs the lowest p63 reactivity in squamous cell carcinoma was 30%.
Interestingly, we noted that p63 was preferentially expressed in TTF-1-positive rather than TTF-1-negative adenocarcinoma (34% vs 10%, P=0.02), whereas CK5/6 and 34βE12 reactivity was independent of TTF-1 (P=0.75 and 0.46, respectively). This explains the high predictive value of p63/TTF-1 combination (ie in majority of cases non-specific p63 in adenocarcinoma is clarified by TTF-1 coexpression).
Analysis of squamous marker coexpression (Table 4) showed that the combinations 100% specific for squamous cell carcinoma were (1) coexpression of p63 and CK5/6 (at any level) in a TTF-1-negative tumor and (2) coexpression of diffuse p63 and diffuse CK5/6 irrespective of TTF-1. This is because p63 and CK5/6 in adenocarcinoma were expressed independently of each other (Spearman correlation coefficient r=−0.02), and coexpression (always focal) occurred in only a small subset of tumors (5%). Of note, a profile that was not seen in any adenocarcinoma or squamous cell carcinoma was diffuse expression of all three markers (TTF-1, p63, CK5/6). The significant overlap in single-marker expression but highly distinct patterns of TTF-1/p63/CK5/6 coexpression profiles in adenocarcinoma vs squamous cell carcinoma can be visualized in a Heatmap (Figure 3).
Heatmap of TTF-1, p63, CK5/6, and 34βE12 coexpression profiles in adenocarcinoma vs squamous cell carcinoma. Individual tumor samples are represented as rows and marker expression as columns. Samples are ordered by decreasing expression of TTF-1, p63, CK5/6, and 34βE12. Expression level (percentage of immunoreactive cells) is indicated by graded color from red (highest) to white (none). ADC, adenocarcinoma; SQCC, squamous cell carcinoma.
Algorithm
Based on the above data we devised an algorithm depicted in Figure 4. This algorithm assumes adenocarcinoma and squamous cell carcinoma as the only diagnostic possibilities, while the differential diagnosis expanded to other tumor types (such as metastases) and caveats specific to small specimens are addressed in the Discussion. In this algorithm, TTF-1 and p63 are used the first-line panel, and CK5/6 is added the third marker in a minority of cases with indeterminate profile. Because diffuse TTF-1 is specific for adenocarcinoma, a further step-wise approach with TTF-1 alone as the first marker (with diffuse reactivity supporting adenocarcinoma vs other reactivities remaining indeterminate) is suggested for cases with extremely limited cellularity.
Data summary and algorithm utilizing TTF-1/p63 as the first-line panel and CK5/6 for indeterminate cases. This algorithm addressed the distinction of adenocarcinoma vs squamous cell carcinoma as the only choices and is based on analysis of whole-tissue sections. The differential diagnosis expanded to other tumor types and caveats specific to small specimens are addressed in the Discussion. (−) 0% reactive cells, (+) <50% reactive cells, (++) ≥50% reactive cells. ADC, adenocarcinoma; SQCC, squamous cell carcinoma; INDET, indeterminate. (1) Although all TTF-1/p63 double-negative cases in resections were adenocarcinoma, there are caveats with applying this profile as a classifier in small specimens (see Discussion); therefore, a favored or indeterminate classification is recommended. (2) All cases with this profile had a high TTF-1/p63 ratio. (3) Focal TTF-1 reactivity in rare squamous cell carcinomas is likely due to lower-specificity TTF-1 clone (SPT24) used in this study (see Discussion). All cases had a low TTF-1/p63 ratio, and diffuse CK5/6.
Because CK5/6 has a lower predictive value than p63 either alone or in combination with TTF-1, it does not change the interpretation of TTF-1/p63 panel outside of the indeterminate setting outlined above. Due to extremely low specificity and no advantage in sensitivity over p63, 34βE12 is not used in the algorithm.
Algorithm Validation in Preoperative Small Biopsy and Cytology Specimens
The above algorithm was prospectively applied to 38 consecutive preoperative small biopsy and cytology specimens of non-small cell carcinoma unclassifiable by morphology, and for which subsequent resection diagnoses was adenocarcinoma or squamous cell carcinoma. As shown in Table 5, this panel was 100% accurate at predicting the tumor type. No TTF-1/p63-indeterminate profiles were encountered in this validation series, and a two-marker panel was sufficient for classification of all specimens. Of note, while immunohistochemistry had excellent accuracy of determining cell lineage as adenocarcinoma vs squamous cell carcinoma, it did not exclude the possibility of other unsampled components in combined carcinomas. On re-review, other components were either absent or not well represented in the preoperative specimens.
Discussion
Several recent studies have evaluated immunohistochemistry and special stain algorithms for non-small cell carcinoma subtyping.14, 15, 16, 17, 23, 24, 25 Because of a large number of cases and use of whole-tissue sections in this study, we were able to examine in greater detail the spectrum of possible coexpression profiles of commonly used markers in adenocarcinoma and squamous cell carcinoma, which serves as the basis for a more fine-tuned algorithm. Specifically, we show that the vast majority of specimens can be classified by TTF-1 and p63, with a third marker (CK5/6) being needed in only a small subset of cases. Whereas most prior studies agree on the inclusion of TTF-1 and p63 in the diagnostic panels, various other markers were included as well, whereas we clarify that TTF-1/p63 panel is sufficient to identify the line of differentiation in the majority of cases based on the proposed algorithm.
Similar to our findings, there is a general agreement in recent studies that classic profiles (TTF-1-diffuse/p63-negative vs TTF-1-negative/p63-diffuse) support the diagnosis of adenocarcinoma vs squamous cell carcinoma, respectively. These profiles are non-overlapping, and are not detected in the opposite tumor type in this or prior studies. In contrast, there remains uncertainty regarding the interpretation of (1) TTF-1/p63 double-positive and (2) TTF-1/p63 double-negative profiles, which is clarified in this study.
Interpretation of TTF-1/p63 double-positive profile: We show conclusively that coexpression of p63 (usually focal but occasionally diffuse) is common in TTF-1-positive adenocarcinoma, whereas diffuse TTF-1 expression is not seen in squamous cell carcinoma. Therefore, coexpression of p63 in a TTF-1 diffusely positive tumor should not be considered a limitation of a panel (as has been suggested24), and such profile should be interpreted as supporting adenocarcinoma. This is in agreement with the conclusion recently reached by several other investigators.16, 17, 29 Of note, it is important to assure that expression of TTF-1 and p63 represents true coexpression (ie reactivity is in the same cell population), whereas expression in separate cell populations may suggest adenosquamous carcinoma.
Surprisingly, we identified that rare squamous cell carcinoma in our series had focal reactivity for TTF-1, whereas most studies suggest that squamous cell carcinoma is consistently TTF-1-negative.16, 17, 29 The reason for this reactivity is most likely the use of a lower-specificity TTF-1 antibody (SPT24; NovoCastra) in this study, whereas it was previously shown that a higher-specificity (but lower sensitivity) TTF-1 clone 8G7G3/1 (DAKO) is consistently non-reactive in squamous cell carcinomas positive by SPT24.30 All squamous cell carcinomas with TTF-1 reactivity in this study were negative for a novel adenocarcinoma marker Napsin A (data not shown) further supporting that this labeling is likely non-specific and is unrelated to glandular differentiation. Although this is a theoretical pitfall for classification of double-positive tumors as adenocarcinoma in laboratories utilizing SPT24 antibody, this is readily resolved by taking into account the ratio of TTF-1/p63 reactivity (low in squamous cell carcinoma vs high in adenocarcinoma) or p63/CK5/6 coexpression in squamous cell carcinoma. Of note, in studies utilizing a more specific TTF-1 clone 8G7G3/1, it was suggested that any reactivity for TTF-1 can be considered diagnostic of adenocarcinoma without quantitation.16, 17
Interpretation of TTF-1/p63 double-negative profile: Another cited limitation of TTF-1/p63 panel is the interpretation of double-negative tumors, which in this study account for 10% of adenocarcinoma and none of squamous cell carcinoma. Our data suggest that double-negative profile excludes squamous cell carcinoma (because p63 is a highly stable marker in squamous cell carcinoma), but it is entirely consistent with adenocarcinoma (because TTF-1-negative adenocarcinoma are not unusual). Several other studies agree with the observation that p63 is consistently positive in pulmonary squamous cell carcinoma,16, 20, 29, 31 and is a highly stable marker irrespective of the grade of differentiation.19 It is possible that rare tumors reported as p63-negative squamous cell carcinoma in some studies represent a solid variant of adenocarcinoma.
While our data support that double-negative profile in whole-tissue sections is consistent with adenocarcinoma but not squamous cell carcinoma, application of this conclusion to small specimens in clinical practice has several caveats. (1) False-negative immunoreactivity that may occur due to technical failure may be difficult to exclude in small specimens where labeling of internal positive control cells (pneumocytes for TTF-1 and bronchial basal cells for p63) may not be represented. (2) Even though p63 is consistently diffuse in squamous cell carcinoma, in extremely small specimens one cannot exclude that the lack of labeling represents a sampling error. (3) Double-negative profile may be seen in various tumor types other than adenocarcinoma (see below). (4) Finally, further study is needed to determine stability of p63 in squamous cell carcinoma with sarcomatoid/pleomorphic features. Therefore, we propose that TTF-1/p63 double-negative profile in small specimens could be interpreted as favoring adenocarcinoma only if positive controls are labeling appropriately, the tumor is well sampled, tumor types other than non-small cell carcinoma are excluded, and there are no sarcomatoid/pleomorphic features. Notably, although the above limitations are important to bear in mind, in recent studies TTF-1/p63 double-negative non-small cell carcinoma in small specimens consistently corresponded to adenocarcinoma (or non-squamous carcinoma) in matched resections,16, 17, 29 similar to our findings.
This study identified that the only TTF-1/p63 profile that overlaps between adenocarcinoma and squamous cell carcinoma is TTF-1-negative/p63-focal (as also recently identified by Mukhopadhyay and Katzenstein16). This profile is seen in only a small subset of both tumor types, and in this series addition of CK5/6 was able to classify these rare cases. However, we recommend that only positive CK5/6 is used as a classifier in this setting (because such coexpression profile is restricted to squamous cell carcinoma), whereas negative CK5/6 is not a reliable discriminator and such cases should retain an unclassified designation.
Although very high predictive value is achieved by TTF-1/p63 panel when the only possibilities are adenocarcinoma and squamous cell carcinoma, interpretation of immunohistochemical profiles in clinical practice requires careful clinicopathologic correlation to exclude possibilities other than non-small cell lung carcinoma. For example, TTF-1-positive tumors include small cell carcinoma of lung and other sites, and carcinomas of thyroid origin. In addition, reactivity for TTF-1 (primarily SPT24 clone) is reported in a subset of several non-lung/non-thyroid carcinomas, including carcinomas of gynecologic tract32 and breast.33 Other than squamous cell carcinoma, p63-positive tumors include metastatic urothelial carcinoma, myoepithelial neoplasms, and metastatic squamous cell carcinoma from other sites. Furthermore, unexpected p63 expression is seen in some non-epithelial tumors, such as lymphoma and sarcoma.18 As mentioned above, the widest and most difficult differential diagnosis is for tumors with TTF-1/p63 double-negative profile, which includes non-epithelial tumors (such as epithelioid melanoma, sarcoma, lymphoma), metastatic carcinoma, and other primary lung tumors (such as mesothelioma). Careful clinicopathologic correlation and, if needed, additional immunostains are essential to exclude these possibilities.
Another area of uncertainty clarified by this study is the interpretation of conflicting CK5/6 reactivity in relation to TTF-1/p63 panel. We find that CK5/6 is much more variable than p63 in squamous cell carcinoma (20% of poorly differentiated squamous cell carcinoma have absent or minimal CK5/6 labeling), and therefore negative CK5/6 does not alter the interpretation of a TTF-1-negative/p63-diffuse profile as squamous cell carcinoma. Similarly, although diffuse CK5/6 is highly unusual in adenocarcinoma, in the absence of p63 it is not sufficient to support squamous differentiation. Overall, we suggest that CK5/6 does not alter the interpretation of typical TTF-1/p63 profiles outside of clarifying the indeterminate setting (TTF-1-negative/p63-focal or equivocal) discussed above.
Similar to other recent studies, our findings emphasize the extremely low specificity of 34βE12,15, 16 even if used in combination with TTF-1. Majority of lung adenocarcinoma label for 34βE12, and nearly half diffusely. Most importantly, TTF-1-negative/34βE12-diffuse profile can occur in adenocarcinoma, and does not serve as a classifier for squamous cell carcinoma. Therefore, the only possible role for 34βE12 is for negative reactivity to serve as evidence against squamous cell carcinoma (although this can be achieved with equivalent sensitivity by p63), whereas positive 34βE12 even if strong and diffuse and present in a TTF-1-negative tumor has no diagnostic value for the distinction of adenocarcinoma vs squamous cell carcinoma. Therefore, inclusion of 34βE12 in the standard panel is not recommended.
A potential limitation of the two-marker algorithm is that some profiles require quantitation of reactivities as focal vs diffuse, which is straightforward in whole-tissue sections but can be a challenge in small specimens. However, we did not find this to present a difficulty in the validation set of small specimens. This is because expression of TTF-1 is bimodal in the majority of adenocarcinoma (either homogenously positive or completely negative) and similarly p63 is consistently diffuse in squamous cell carcinoma, and such reactivities are readily distinguishable from uncommon focal reactivities even in small specimens.
Several other markers have been reported to be useful in differentiating adenocarcinoma from squamous cell carcinoma. In particular, recent studies show that a relatively new adenocarcinoma marker Napsin A can identify a subset of TTF-1-negative adenocarcinoma,34 whereas this marker is consistently negative in squamous cell carcinoma.16, 17 Therefore, Napsin A may be useful to further support the interpretation of a double-negative (or TTF-1-equivocal) profile as adenocarcinoma. Mucin stains (mucicarmine, PAS) have been the markers of choice for the diagnosis of adenocarcinoma in preimmunohistochemistry era, but their sensitivity for adenocarcinoma is low (23–30%).14, 25 Another frequently cited marker for adenocarcinoma is CK7, but it is also positive in a substantial proportion (30–60%) of pulmonary squamous cell carcinoma, particularly peripheral squamous cell carcinoma.16, 35 In addition, several new squamous markers (Desmocollin-3,36 microRNA miR-205,29 Sox2,37 and S100A738) have been recently described as useful for non-small cell carcinoma subtyping. Overall, given the high accuracy that can be achieved by widely available TTF-1/p63 panel, it is important to determine whether there is an added value provided by these additional markers. In fact, recently it was shown that miR-205 is inferior to immunohistochemistry for distinguishing adenocarcinoma and squamous cell carcinoma.39
Rather than a step-wise approach proposed here, an alternative method of tissue conservations is the use of antibody cocktails, as recently described.17, 40 While promising, the wide utilization may be limited by availability of these reagents. In contrast, the algorithm described here is widely applicable because it includes conventional well-characterized antibodies, which are available in most pathology laboratories. The fact that TTF-1 and p63 are nuclear markers is also advantageous because interpretation is less often complicated by artifactual or equivocal staining that can occur with cytoplasmic markers. Finally, these findings are biologically sensible as it is becoming recognized that TTF-1 and p63 are master regulators of differentiation in glandular28 and squamous21, 41 cell lineages, respectively.
Whereas immunohistochemistry is emerging as a powerful tool in non-small cell carcinoma subtyping, it is important to emphasize that the majority of adenocarcinoma and squamous cell carcinoma can be distinguished based on standard morphologic criteria. This is illustrated in a recent review of clinical practice at our institution, showing that 88% of cytologic specimens with subsequent resection diagnosis of adenocarcinoma and squamous cell carcinoma were subtyped without the utilization of immunostains.18 Reviews from several other institutions also show that of the majority small biopsy/cytology specimens of non-small cell carcinoma are classifiable morphologically, and immunostains are needed in only a subset of cases.8, 14, 17
Although the primary goal of this study was to devise an algorithm for non-small cell carcinoma subtyping in small specimens, the results also have potential implications for large cell carcinoma—a tumor defined in resected specimens as unclassifiable non-small cell carcinoma with no apparent glandular or squamous differentiation by morphology.7 Several recent studies showed that immunostains (and previously electron microscopy42) can reveal the line of differentiation as adenocarcinoma vs squamous cell carcinoma in the majority of large cell carcinomas.26, 36, 43, 44 The proposed TTF-1/p63 algorithm could be used to determine biological identity within the large cell carcinoma, but this needs further study.
The biological significance of ‘squamous marker’ expression in lung adenocarcinoma is currently unknown. Preferential expression of p63 in TTF-1-positive adenocarcinoma identified in this study is intriguing, as is the recent finding of preferential p63 expression in adenocarcinoma with EML4-ALK translocations,45 suggesting that this expression may not be simply aberrant. While further studies are needed to address the biological role, what can be concluded at this time is that ‘squamous marker’ expression is common in lung adenocarcinoma, and it presents a potential diagnostic pitfall if interpreted in the absence of TTF-1. Furthermore, findings so far suggest that this expression does not indicate coordinate activation of squamous differentiation program because (1) several markers are expressed independently of each other and (2) there is no association with squamous morphology (unpublished data). Therefore, isolated detection of p63, CK5/6, or 34βE12 in adenocarcinoma in the absence of overt squamous morphology should not be interpreted as evidence for adenosquamous carcinoma.
In summary, we used a large number of whole-tissue sections to examine in detail coexpression profiles of four markers commonly used to distinguish adenocarcinoma and squamous cell carcinoma, and prospectively validated the devised algorithm in a series of small specimens. We find that a two-marker panel of TTF-1/p63 is sufficient for subtyping of the majority of adenocarcinoma and squamous cell carcinoma, and addition of CK5/6 (and Napsin A, based on other studies) is useful in only a subset of cases. Whereas performing an expanded four-marker panel (TTF-1, Napsin A, p63, CK5/6) upfront is a time-efficient option for specimens with sufficient cellularity, the proposed step-wise approach allows prioritization of markers if the amount of tissue or resources is limited, and yields optimal conservation of tissue for predictive marker testing.
References
Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst 2005;97:339–346.
Johnson DH, Fehrenbacher L, Novotny WF, et al. Randomized phase II trial comparing bevacizumab plus carboplatin and paclitaxel with carboplatin and paclitaxel alone in previously untreated locally advanced or metastatic non-small-cell lung cancer. J Clin Oncol 2004;22:2184–2191.
Scagliotti G, Brodowicz T, Shepherd FA, et al. Treatment-by-histology interaction analyses in three phase III trials show superiority of pemetrexed in nonsquamous non-small cell lung cancer. J Thorac Oncol 2011;6:64–70.
Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol 2009;27:4247–4253.
Bass AJ, Watanabe H, Mermel CH, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet 2009;41:1238–1242.
Weiss J, Sos ML, Seidel D, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med 2010;2:62ra93.
Travis WD, Brambilla E, Muller-Hermelink HK, Harris CC, (eds). World Health Organization Classification of Tumours. Pathology & Genetics: Tumours of the Lung, Pleura, Thymus and Heart. IARC Press: Lyon, 2004.
Edwards SL, Roberts C, McKean ME, et al. Preoperative histological classification of primary lung cancer: accuracy of diagnosis and use of the non-small cell category. J Clin Pathol 2000;53:537–540.
Ou SH, Zell JA . Carcinoma NOS is a common histologic diagnosis and is increasing in proportion among non-small cell lung cancer histologies. J Thorac Oncol 2009;4:1202–1211.
Travis WD, Rekhtman N, Riley GJ, et al. Pathologic diagnosis of advanced lung cancer based on small biopsies and cytology: a paradigm shift. J Thorac Oncol 2010;5:411–414.
Rossi G, Pelosi G, Graziano P, Barbareschi M, Papotti M . A reevaluation of the clinical significance of histological subtyping of non--small-cell lung carcinoma: diagnostic algorithms in the era of personalized treatments. Int J Surg Pathol 2009;17:206–218.
Travis WD, Brambilla E, Noguchi M, et al. International association for the study of lung cancer/American thoracic society/European respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 2011;6:244–285.
Rossi G, Papotti M, Barbareschi M, Graziano P, Pelosi G . Morphology and a limited number of immunohistochemical markers may efficiently subtype non-small-cell lung cancer. J Clin Oncol 2009;27:e141–e142; author reply e3–e4.
Loo PS, Thomas SC, Nicolson MC, Fyfe MN, Kerr KM . Subtyping of undifferentiated non-small cell carcinomas in bronchial biopsy specimens. J Thorac Oncol 2010;5:442–447.
Nicholson AG, Gonzalez D, Shah P, et al. Refining the diagnosis and EGFR status of non-small cell lung carcinoma in biopsy and cytologic material, using a panel of mucin staining, TTF-1, cytokeratin 5/6, and P63, and EGFR mutation analysis. J Thorac Oncol 2010;5:436–441.
Mukhopadhyay S, Katzenstein AL . Subclassification of non-small cell lung carcinomas lacking morphologic differentiation on biopsy specimens: utility of an immunohistochemical panel containing TTF-1, Napsin A, p63, and CK5/6. Am J Surg Pathol 2011;35:15–25.
Righi L, Graziano P, Fornari A, et al. Immunohistochemical subtyping of nonsmall cell lung cancer not otherwise specified in fine-needle aspiration cytology: a retrospective study of 103 cases with surgical correlation. Cancer 2011; e-pub ahead of print.
Rekhtman N, Brandt SM, Sigel CS, et al. Suitability of thoracic cytology for new therapeutic paradigms in non-small cell lung carcinoma: high accuracy of tumor subtyping and feasibility of EGFR and KRAS molecular testing. J Thorac Oncol 2011;6:451–458.
Wang BY, Gil J, Kaufman D, et al. P63 in pulmonary epithelium, pulmonary squamous neoplasms, and other pulmonary tumors. Hum Pathol 2002;33:921–926.
Au NH, Gown AM, Cheang M, et al. P63 expression in lung carcinoma: a tissue microarray study of 408 cases. Appl Immunohistochem Mol Morphol 2004;12:240–247.
Pelosi G, Pasini F, Olsen Stenholm C, et al. p63 immunoreactivity in lung cancer: yet another player in the development of squamous cell carcinomas? J Pathol 2002;198:100–109.
Camilo R, Capelozzi VL, Siqueira SA, Del Carlo Bernardi F . Expression of p63, keratin 5/6, keratin 7, and surfactant-A in non-small cell lung carcinomas. Hum Pathol 2006;37:542–546.
Khayyata S, Yun S, Pasha T, et al. Value of P63 and CK5/6 in distinguishing squamous cell carcinoma from adenocarcinoma in lung fine-needle aspiration specimens. Diagn Cytopathol 2009;37:178–183.
Ring BZ, Seitz RS, Beck RA, et al. A novel five-antibody immunohistochemical test for subclassification of lung carcinoma. Mod Pathol 2009;22:1032–1043.
Terry J, Leung S, Laskin J, et al. Optimal immunohistochemical markers for distinguishing lung adenocarcinomas from squamous cell carcinomas in small tumor samples. Am J Surg Pathol 2010;34:1805–1811.
Conde E, Angulo B, Redondo P, et al. The use of P63 immunohistochemistry for the identification of squamous cell carcinoma of the lung. PLoS One 2010;5:e12209.
Travis WD, Rekhtman N . Pathologic diagnosis and classification of lung cancer in small biopsies and cytology: the role of special stains and need for strategic management of tissue for molecular testing. Semin Respir Crit Care Med 2011;32:22–31.
Yatabe Y, Mitsudomi T, Takahashi T . TTF-1 expression in pulmonary adenocarcinomas. Am J Surg Pathol 2002;26:767–773.
Bishop JA, Benjamin H, Cholakh H, et al. Accurate classification of non-small cell lung carcinoma using a novel microRNA-based approach. Clin Cancer Res 2010;16:610–619.
Matoso A, Singh K, Jacob R, et al. Comparison of thyroid transcription factor-1 expression by 2 monoclonal antibodies in pulmonary and nonpulmonary primary tumors. Appl Immunohistochem Mol Morphol 2010;18:142–149.
Moreira AL, Gonen M, Rekhtman N, Downey RJ . Progenitor stem cell marker expression by pulmonary carcinomas. Mod Pathol 2010;23:889–895.
Zhang PJ, Gao HG, Pasha TL, Litzky L, Livolsi VA . TTF-1 expression in ovarian and uterine epithelial neoplasia and its potential significance, an immunohistochemical assessment with multiple monoclonal antibodies and different secondary detection systems. Int J Gynecol Pathol 2009;28:10–18.
Robens J, Goldstein L, Gown AM, Schnitt SJ . Thyroid transcription factor-1 expression in breast carcinomas. Am J Surg Pathol 2010;34:1881–1885.
Bishop JA, Sharma R, Illei PB . Napsin A and thyroid transcription factor-1 expression in carcinomas of the lung, breast, pancreas, colon, kidney, thyroid, and malignant mesothelioma. Hum Pathol 2009;41:20–25.
Saijo T, Ishii G, Nagai K, et al. Differences in clinicopathological and biological features between central-type and peripheral-type squamous cell carcinoma of the lung. Lung Cancer 2006;52:37–45.
Monica V, Ceppi P, Righi L, et al. Desmocollin-3: a new marker of squamous differentiation in undifferentiated large-cell carcinoma of the lung. Mod Pathol 2009;22:709–717.
Yuan P, Kadara H, Behrens C, et al. Sex determining region Y-Box 2 (SOX2) is a potential cell-lineage gene highly expressed in the pathogenesis of squamous cell carcinomas of the lung. PLoS One 2010;5:e9112.
Zhang H, Zhao Q, Chen Y, et al. Selective expression of S100A7 in lung squamous cell carcinomas and large cell carcinomas but not in adenocarcinomas and small cell carcinomas. Thorax 2008;63:352–359.
Del Vescovo V, Cantaloni C, Cucino A, et al. miR-205 expression levels in nonsmall cell lung cancer do not always distinguish adenocarcinomas from squamous cell carcinomas. Am J Surg Pathol 2011;35:268–275.
Yanagita E, Imagawa N, Ohbayashi C, Itoh T . Rapid multiplex immunohistochemistry using the 4-antibody cocktail YANA-4 in differentiating primary adenocarcinoma from squamous cell carcinoma of the lung. Appl Immunohistochem Mol Morphol 2011; e-pub ahead of print.
Hibi K, Trink B, Patturajan M, et al. AIS is an oncogene amplified in squamous cell carcinoma. Proc Natl Acad Sci USA 2000;97:5462–5467.
Churg A . The fine structure of large cell undifferentiated carcinoma of the lung. Evidence for its relation to squamous cell carcinomas and adenocarcinomas. Hum Pathol 1978;9:143–156.
Rossi G, Marchioni A, Milani M, et al. TTF-1, cytokeratin 7, 34betaE12, and CD56/NCAM immunostaining in the subclassification of large cell carcinomas of the lung. Am J Clin Pathol 2004;122:884–893.
Pardo J, Martinez-Penuela AM, Sola JJ, et al. Large cell carcinoma of the lung: an endangered species? Appl Immunohistochem Mol Morphol 2009;17:383–392.
Yoshida A, Tsuta K, Watanabe SI, et al. Frequent ALK rearrangement and TTF-1/p63 co-expression in lung adenocarcinoma with signet-ring cell component. Lung Cancer 2010;72:309–315.
Acknowledgements
We thank Marina Asher and Irina Linkoff for performing immunohistochemistry.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Rekhtman, N., Ang, D., Sima, C. et al. Immunohistochemical algorithm for differentiation of lung adenocarcinoma and squamous cell carcinoma based on large series of whole-tissue sections with validation in small specimens. Mod Pathol 24, 1348–1359 (2011). https://doi.org/10.1038/modpathol.2011.92
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2011.92
Keywords
This article is cited by
-
Validity of using immunohistochemistry to predict treatment outcome in patients with non-small cell lung cancer not otherwise specified
Journal of Cancer Research and Clinical Oncology (2019)
-
A lung cancer risk classifier comprising genome maintenance genes measured in normal bronchial epithelial cells
BMC Cancer (2017)