Abstract
Ovarian tumors of low malignant potential and low-grade ovarian serous carcinomas are thought to represent different stages on a tumorigenic continuum and to develop along pathways distinct from high-grade ovarian serous carcinoma. We performed gene expression profiling on three normal human ovarian surface epithelia samples, and 10 low-grade and 10 high-grade ovarian serous carcinomas. Analysis of gene expression profiles of these samples has identified 80 genes upregulated and 232 genes downregulated in low-grade ovarian serous carcinomas. PAX2 was found to be one of the most upregulated genes in low-grade ovarian serous carcinoma. The upregulation of PAX2 was validated by real-time quantitative RT-PCR, western blot and immunohistochemical analyses. Real-time RT-PCR showed a statistically significant difference in PAX2 mRNA expression (expressed as fold change in comparison to normal human ovarian surface epithelia) among ovarian tumors of low malignant potential (1837.38, N=8), low-grade (183.12, N=17), and high-grade (3.72, N=23) carcinoma samples (P=0.015). Western blot analysis revealed strong PAX2 expression in ovarian tumors of low malignant potential (67%, N=3) and low-grade carcinoma samples (50%, N=10) but no PAX2 protein expression in high-grade carcinomas (0%, N=10). Using immunohistochemistry, tumors of low malignant potential (59%, N=17) and low-grade carcinoma (63%, N=16) samples expressed significantly stronger nuclear staining than high-grade ovarian carcinoma samples (9.1%, N=263). Furthermore, consistent with earlier immunohistochemical findings, PAX2 expression was expressed in the epithelial cells of fallopian tubes but not in normal ovarian surface epithelial cells. Our findings further support the two-tiered hypothesis that tumors of low malignant potential and low-grade ovarian serous carcinoma are on a continuum and are distinct from high-grade ovarian carcinomas. In addition, the absence of PAX2 expression in normal ovarian epithelia but expression in fallopian tube fimbria and ciliated epithelial inclusions would suggest the potential development of tumors of low malignant potential and of low-grade ovarian serous carcinomas from secondary Müllerian structures.
Similar content being viewed by others
Main
Ovarian cancer, a leading cause of cancer deaths among women in the United States, is estimated to affect over 20 000 women each year, with over 15 000 dying of the disease.1 Serous carcinoma, the major histologic subtype of ovarian cancer, represents about 87% of advanced-staged cases.2 Currently, no uniform grading system exists despite the fact that histologic grade is a significant prognostic factor in women with epithelial ovarian carcinoma.3 Malpica et al proposed a two-tiered grading system for ovarian serous carcinoma based on nuclear atypia and mitotic rate and classified tumors into low grade vs high grade.4 Low-grade carcinomas and ovarian serous tumors of low malignant potential are often associated together in histologic and molecular studies and are rarely linked with high-grade ovarian carcinoma.4, 5 Differences in clinical behavior further support the distinction between the two grades. Low-grade carcinomas are typically characterized by a younger patient population with longer overall survival and increased platinum chemoresistance compared with high-grade carcinomas.6 These variations in histologic and clinical behavior have led investigators to suggest that low-grade and high-grade ovarian serous carcinomas may develop along different pathways, and tumors of low malignant potential may exist on a continuum with low-grade ovarian serous carcinoma.
The precise etiology and pathogenesis of ovarian carcinoma remains elusive despite significant research. The cell of origin is still widely unknown and debated. The most common hypothesis is that ovarian carcinomas develop from the surface epithelium or postovulatory inclusion cysts in response to trauma or prolonged exposure to hormones or other chemokines.7, 8, 9 However, growing evidence suggests other possible etiologies, including the tubal epithelium and distal endosalpinx.10, 11 Molecular and translational studies have suggested that differences in development pathways may contribute to the clinical, histologic, and genetic differences between low-grade and high-grade ovarian serous carcinomas.12
Further evidence is needed, however, to establish definitively that ovarian tumors of low malignant potential progress to low-grade ovarian serous carcinomas and that the developmental pathway of these two is distinct from that of high-grade ovarian serous carcinomas. Toward that end, in this study, we utilized gene expression profiling to identify a variety of genes that are differentially expressed in tumors of low malignant potential and low-grade ovarian serous carcinomas compared with high-grade ovarian serous carcinomas. From the list of differentially expressed genes, we identified PAX2 to be highly upregulated in low-grade ovarian carcinomas. Thus, we further validated the expression of PAX2 at the mRNA and protein level and demonstrated its significantly higher expression in tumors of low malignant potential and low-grade ovarian serous carcinomas than in high-grade ovarian serous carcinomas. The differential PAX2 expression in tumors of low malignant potential and low-grade tumors supports the two-tiered system of serous ovarian cancer grading and endorses further potentially different development pathways in low- and high-grade ovarian serous carcinoma.
Materials and methods
Human Subjects and Tissue Specimens
Tissue specimens were obtained and stored in accordance with the human subject research protocols approved by the institutional review board. Frozen tissue sections of tumors of low malignant potential and of low-grade and high-grade carcinomas were obtained from the tumor repository in the Department of Gynecologic Oncology at The University of Texas MD Anderson Cancer Center. Lysates were obtained from these bulk tumors, and pathologic review of frozen sections showed at minimum a 70% tumor cell component. Sections from three human ovarian surface epithelia and from 10 low-grade and 10 high-grade ovarian serous carcinoma samples were utilized to perform gene expression profiling. We also examined levels of PAX2 expression from total RNA extracted from a second independent frozen tissue section set consisting of eight low malignant potential tumors and 17 low-grade and 23 high-grade ovarian carcinomas. A third independent set of paraffin tissue samples (two normal human ovarian surface epithelia, three normal fallopian tubes, 17 low malignant potential tumors, 16 low-grade and 263 high-grade carcinomas) from the University of Texas MD Anderson Cancer Center were also obtained and used for immunohistochemistry (IHC) validation studies. All tissue samples were reviewed and examined by specialized gynecologic oncology pathologists and graded according to criteria outlined by Malpica et al.4
Expression Microarray
Frozen tissue samples of 10 low-grade and 10 high-grade ovarian serous carcinomas containing >70% tumor cells were homogenized in preparation for RNA extraction. RNA was obtained from short-term cultures of three different normal human ovarian surface epithelial cells that were initiated from the surface scraping of normal ovaries before their removal from patients with nonmalignant diseases. RNA extraction was performed using the Mini-prep RNeasy kit (Qiagen, Valencia, CA, USA) per the manufacturer's protocol. Expression profiles were generated by using Affymetrix Human Genome U133 plus 2.0 (HG U133 plus 2.0) Gene Chips (Affymetrix, Santa Clara, CA, USA) according to the Affymetrix Eukaryotic One-cycle protocol. Briefly, 5 μg of total RNA was used to generate biotinylated cRNA, which was then fragmented by heat and ion-mediated hydrolysis at 94°C for 35 min in 24 μl H2O and 6 μl of 5 × fragmentation buffer (Affymetrix). The fragmented cDNA was hybridized to the HG U133 plus 2.0 Gene Chip (Affymetrix) for 16 h at 45°C in an Affymetrix Hybridization Oven 640 (Affymetrix). Arrays were then washed and stained on an Affymetrix Fluidics Station 450 (Affymetrix) and subsequently scanned on an Affymetrix GeneChip Scanner 3000 and GeneChip Operating Software (Affymetrix) to obtain fluorescence intensities. The resulting raw images (CEL files) were processed with dChip software,13 and unsupervised cluster analysis was performed, as previously described.14
Real-Time RT-PCR
To perform real-time RT-PCR, we obtained and homogenized a second independent set of frozen ovarian tissue sections from eight low malignant potential serous tumors and 17 low-grade and 23 high-grade ovarian serous carcinomas. Again, RNA extraction using the Mini-prep RNeasy kit (Qiagen) was performed. cDNA synthesis was constructed from samples of all three normal human ovarian surface epithelia, eight low malignant potential, 17 low-grade, and 23 high-grade RNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA). A 25-μl solution containing 1 μg RNA, 10 × RT buffer, 25 × dNTPs, and 10 × random hexamers was created for each RNA sample. cDNA amplification was done for 1 cycle at 25°C for 10 min, at 37°C for 60 min, and at 95°C for 5 min. The reaction was stopped using 1 μl of 50 mM EDTA and diluted 1:10 using sterile dH2O. Each real-time RT-PCR was performed in triplicate. For each reaction, 5 μl of each cDNA was combined with 10 μl 2 × Brilliant II QPCR Master Mix (Stratagene, La Jolle, CA, USA), 3.7 μl dH2O, 0.3 μl 1:500 diluted reference dye, and 1 μl 20 × PAX2 Taqman gene expression assay mix (Applied Biosystems). All results were normalized to cyclophilin A pre-developed Taqman expression gene assay (Applied Biosystems). mRNA quantification was determined using MX4000 Multiplex Quantification PCR System (Stratagene) for 1 cycle at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The mRNA copy number for tumor samples was compared with that for samples of normal ovarian surface epithelial cells.
Western Blot
RIPA buffer (Sigma, St Louis, MO, USA) was added to homogenized samples from three serous ovarian tumors of low malignant potential and 10 low-grade and 10 high-grade ovarian serous carcinomas for protein extraction. Protein concentration was determined using a Beckman DU640B spectrophotometer (Beckman, Fullerton, CA, USA). About 50 μg of denatured protein was loaded and electrophoresed onto a 10% SDS-PAGE gel (Bio-Rad, Hercules, CA, USA), electroblotted on Hybond ECL nitrocellulose membrane (GE Healthcare, Buckinghamshire, UK), and probed using rabbit polyclonal anti-PAX2 antibody (Zymed Laboratories, San Francisco, CA, USA) at a 1:500 dilution. Goat anti-rabbit IgG-HRP (Santa Cruz Biotechnology, Santa Cruz, CA, USA) at a 1:10 000 dilution was used to bind to the anti-PAX2 antibody. The bound antibodies were detected using an Amersham ECL Western Blot Detection reagent kit (GE Healthcare).
Immunohistochemistry
PAX2 expression was validated using an independent set of paraffin tissue sections from The University of Texas MD Anderson Cancer Center. Immunohistochemistry was performed on deparaffinized sections from two normal ovarian epithelia, three normal fallopian tubes, three endosalpingiosis samples from the ovaries, 17 low malignant potential tumors and 16 low-grade and 106 high-grade ovarian serous carcinoma samples. Additional IHC staining of high-grade ovarian serous carcinoma samples was performed using three pre-constructed tissue microarray paraffin blocks (0.6 mm per core) that contained 157 high-grade ovarian carcinoma samples. For antigen retrieval, deparaffinized specimens were immersed in 1 × Reveal (Biocare Medical, Concord, CA, USA) in a Coplin jar and transferred to a Reveal decloaking chamber (Biocare Medical) heated at 121°C for 3 min followed by 1 min at 95°C. Rabbit polyclonal anti-PAX2 antibody (Zymed Laboratories) at a 1:200 dilution was then placed onto the tissue sections at room temperature for 4 h and probed with MACH 3 rabbit probe (Biocare Medical) and MACH 3 rabbit alkaline-phosphatase polymer (Biocare Medical). The tissue sections were subsequently stained using Vulcan Fast Red Chromagen Kit 2 (Biocare Medical) and counterstained using CAT Hematoxylin (Biocare Medical).
Each paraffin section was examined for nuclear staining and cytoplasmic staining for PAX2. However, only nuclear staining was considered indicative of PAX2 expression given that PAX2 encodes a transcription factor associated with organogenesis. Nuclear staining intensity was graded as negative, weak, moderate, or strong. The percentage of positive cells for each sample was also noted.
Statistical Analysis
Statistical analyses were performed using SPSS 15.0 software (SPSS, Chicago, IL, USA). Kruskall–Wallis and Mann–Whitney U nonparametric tests were utilized to compare differences between PAX2 expression levels. P-values of <0.05 were considered statistically significant.
Results
Identification of Differential PAX2 Gene Expression
The expression profiles of the samples in this study were deposited at the Gene Expression Omnibus public database with access number GSE14001. Unsupervised clustering analysis of these samples with a list of 5131 genes with coefficient of variation in expression level between 0.5 and 1000 was performed. Figure 1 indicates that low-grade ovarian serous carcinoma and high-grade ovarian serous carcinoma clustered into two different groups except sample HG01-161 which clustered with low-grade ovarian carcinoma. To identify differentially expressed genes between low-grade and high-grade ovarian serous carcinoma, statistical analysis was performed by SAM (significance analysis of microarrays) software, which is a t-test analysis with permutation to estimate false discovery. Eighty genes were found to be upregulated in low-grade ovarian serous carcinoma and 232 genes were found to be downregulated in low-grade ovarian serous carcinoma with more than twofold with a false discovery of less than 20% (Supplementary Table 1). Four of the most differentially expressed genes were further validated by real-time RT-PCR (Table 1).
We selected PAX2 for further validation because it has been demonstrated to have high expression throughout the Mullerian duct system and to be involved in the development of breast and urogenital cancers.10, 15, 16 In our study, PAX2 expression in low-grade ovarian serous carcinomas showed one of the highest fold-increases compared with its expression in the high-grade carcinomas or in normal human ovarian epithelia. The fold change was statistically significant (mean PAX2 expression 291.19 for low-grade ovarian carcinomas vs 60.89 for high-grade ovarian carcinomas; P=0.040).
PAX2 mRNA Quantification
We used real-time RT-PCR quantification of PAX2 mRNA to validate the expression array data. PAX2 mRNA expression levels differed significantly among ovarian low malignant potential tumor and low-grade and high-grade ovarian carcinoma samples (Figure 2). Although the differences in median PAX2 mRNA expression levels between low malignant potential tumors and low-grade ovarian serous carcinomas were not statistically significant (1837.38 (mean, 3602.4) vs 183.12 (mean, 442.8), respectively (P=0.086)), the median mRNA expression of PAX2 was significantly lower in the high-grade ovarian serous carcinomas (median, 3.72; mean, 148.3) than in either the ovarian low malignant potential tumors or the low-grade ovarian carcinomas (P=0.015).
Comparison of PAX2 mRNA expression between eight tumors of low malignant potential and 17 low-grade and 23 high-grade ovarian carcinoma samples. The box is bounded by the 25th and 75th percentile with the median expression level depicted by the line in the box. Outlying values are drawn individually. Expression of PAX2 in high-grade is significantly lower than either low malignant potential or low-grade tumors (P=0.015).
PAX2 Protein was Expressed in the Nuclei of Low Malignant Potential Tumors and Low-Grade Ovarian Serous Carcinomas
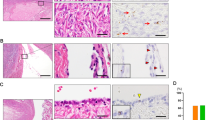
Using western blot analysis, we evaluated PAX2 protein expression in three serous ovarian low malignant potential tumors, 10 low-grade and 10 high-grade serous carcinomas. Sixty-seven percent of low malignant potential tumors (2/3) and 50% of low-grade samples (5/10) demonstrated strong PAX2 expression. Protein extracted from high-grade ovarian carcinomas showed no evidence of PAX2 expression (Figure 3). Figure 3 only showed the part of the western blot result of two low malignant potential tumors, seven low-grade, and seven high-grade ovarian serous carcinoma samples. The specificity of the PAX2 antibody to detect PAX2 protein on the western blot was confirmed on a western blot analysis of a cell line expressing PAX2 protein and a PAX2 knockdown of the same cell line (data not shown). Protein expression results were further validated using IHC studies with a large sample size. Overall, tumor sections were found either to lack nuclear staining or to have strong nuclear staining. No sections expressed weak or moderate strength staining. The number of PAX2-positive tumor cells varied from 1–100%. Of all low malignant potential tumor sections, 12, 29, and 18% had between 1–25, 25–50, and 75–100% positivity, respectively. Within the low-grade carcinoma sections, 13, 25, and 25% had 1–25, 25–50, and 75–100%, respectively. Two, five, and three percent of the 263 high-grade carcinomas had ≤1, 10–50, and 75–100% PAX2 nuclear staining expressed. Ovarian low malignant potential tumors that stained positive for PAX2 expressed robust nuclear staining, as did sections of low-grade ovarian carcinomas (Figure 4). Generally, most high-grade ovarian carcinomas showed no nuclear staining or only had extremely faint cytoplasmic expression. The low malignant potential tumors and low-grade carcinomas had significantly more sections that expressed strong nuclear staining than high-grade carcinomas (P<0.001) (Table 2). In addition, strong, robust PAX2 staining was noted in all sections of normal fallopian tube and ciliated epithelial inclusions, but absent in normal ovarian surface epithelia and benign ovarian cysts (Figure 5).
(A) Examples of PAX2 immunohistochemistry staining of individual paraffin sections from low malignant potential tumors and low-grade and high-grade ovarian serous carcinomas ( × 200 magnification). (B) PAX2 immunohistochemistry staining of high-grade ovarian carcinoma paraffin tissue array sections. (a) × 200 magnification. (b) × 40 magnification. (c) × 200 magnification.
PAX2 immunohistochemistry of (a) normal human ovarian surface epithelial and benign ovarian cyst (BC) which have no PAX2 staining ( × 100 magnification); (b) fallopian tube fimbria (FF) demonstrating robust staining ( × 200 magnification) and (c). Ciliated epithelial inclusion (CEI) demonstrating robust staining ( × 100 magnification).
Discussion
Molecular differences between low-grade and high-grade ovarian carcinomas and their association with ovarian tumors of low malignant potential have been examined in a few previous studies.17, 18, 19, 20 High-grade ovarian serous carcinomas were demonstrated to have p53 mutations and higher expression of bcl-2 and c-kit than low-grade carcinomas, which in many cases have BRAF and KRAS mutations.18, 21 Also, significantly higher levels of estrogen receptor, progesterone receptor, and E-cadherin were found in ovarian low-grade carcinomas than in high-grade carcinomas, as reported in one study.21 Moreover, low malignant potential tumors were found to coexist in 60% of ovarian low-grade ovarian serous carcinomas but in only 2% of high-grade carcinomas,4 and approximately 70–80% of patients with ovarian tumors of low malignant potential who have a relapse, the disease will recur as low-grade ovarian carcinoma.22, 23, 24
In this study, we used gene expression profiling to provide further genetic and molecular evidence to differentiate tumors of low malignant potential and low-grade from high-grade tumors by unsupervised clustering analysis, and have identified a list of differentially expressed genes. The expression of the four most differentially expressed genes, PAX2, SOSTDC1, BST2, and ISG15, were further validated by real-time quantitative RT-PCR using the original RNA samples. The role of PAX2 in ovarian cancer is unknown. Recently, Tong et al identified PAX2 expression in secondary Müllerian structures in the ovary as well as epithelial cells of the fallopian tube but not in ovarian surface epithelium or surface epithelium-derived inclusion cysts by immunohistochemistry.10 They found that 21 out of 30 high-grade ovarian carcinomas were positive for PAX2 staining unlike our low incidence of PAX2 expression in high-grade ovarian carcinoma. Thus, we further validated the expression of PAX2 at the mRNA and protein levels and demonstrated its significantly higher expression in ovarian tumors of low malignant potential and low-grade ovarian serous carcinomas than in high-grade carcinomas. We were not able to detect the PAX2 protein in any of the 10 high-grade ovarian carcinoma protein lysates by western blot. Furthermore, immunohistochemical analysis of 263 high-grade ovarian carcinoma samples (either from individual slides or tissue arrays) detected only 24 high-grade ovarian serous carcinoma samples with positive staining for PAX2. This discrepancy may be because of the difference in the number of samples being tested.
PAX2 is a member of the paired-box (PAX) gene family, which consists of nine genes that have been subclassified into four groups (PAX1/PAX9, PAX 2/PAX5/PAX8, PAX4/PAX6, and PAX 3/PAX7) based on genomic structure, sequence similarities, and expression patterns in developing tissues and organs.25 Each PAX gene encodes a protein that is hypothesized to modify downstream gene transcription by binding to enhancer DNA sequences. These transcription factors regulate tissue development and aid with cell-lineage specification, proliferation, migration, and survival.26 The structural variations between the various PAX transcription factors relate to a conserved paired-box DNA-binding domain that may or may not have an associated octapeptide or homeodomain. PAX genes influence embryogenesis, and expression levels usually attenuate during terminal differentiation of most organ systems.25, 26
PAX2 specifically encodes a transcription factor crucial to the organogenesis and development of the central nervous system, eyes, ears, mammary glands, and urogenital tract.10, 15, 27 Mutations in PAX2 have been associated with renal-coloboma syndrome, and unattenuated PAX2 expression has been found in various cancers, including ovarian, renal, prostate, and breast cancers, Wilms tumor, and Kaposi sarcoma.15, 16, 28, 29, 30, 31 The significance of PAX2 in renal development and in the etiology of renal disease, such as renal cell carcinoma and nephrogenic adenomas, has been widely studied.16, 27, 32
As PAX2 expression may indicate cell-lineage, our findings would suggest that tumors of low malignant potential and low-grade carcinomas may not originate from the ovarian surface epithelia or inclusion cysts, as traditionally believed, but from secondary Müllerian epithelium. Expression of PAX2 in fallopian tube fimbria and ciliated epithelial inclusions but not in normal ovarian surface epithelia supports the possibility that low malignant potential tumor arises from ciliated epithelial inclusion. The differential PAX2 expression also supports the hypothesis that tumors of low malignant potential and low-grade carcinoma exist along a disease continuum and that some low-grade carcinoma cases may arise from low malignant potential tumors. The expression of PAX2 in a limited number of high-grade samples suggests that some high-grade cases may also develop from low malignant potential and low-grade lineage, but the majority of these cases may originate along a different development path.
Increased PAX2 expression levels might explain the decreased proliferative growth and increased chemoresistance of low-grade ovarian carcinoma. The signaling pathway for PAX2 is currently unknown. PAX2 is located on chromosome 10q24 and is associated with cell survival and self-sufficiency.26 Studies have found that inhibition of PAX2 expression decreases proliferation of renal cancer cells.16, 33 In addition, Muratovska et al downregulated PAX2 expression in ovarian cancer cell lines treated with PAX2 small inhibitory double-stranded RNA (siRNA) and demonstrated a decrease in cell proliferation in vitro, further suggesting a function of PAX2 in inhibiting cell apoptosis.34 Furthermore, deregulated PAX2 function has been found to block caspase-2 associated apoptosis.32 PAX2 overexpression has also been implicated in the resistance of renal cell carcinoma to cisplatin-induced apoptosis, and inhibition of PAX2 resulted in increased cisplatin sensitivity.35 Our identification of increased PAX2 expression in low-grade ovarian serous carcinoma may also explain the relative chemoresistance of these carcinomas to traditional chemotherapy. The differences in treatment response and chemosensitivity between low- and high-grade ovarian carcinomas indicate that treatment may need to be individualized based on tumor grade. Chemosensitivity of low-grade tumors to current chemotherapeutic agents may improve with the development of targeted therapies to PAX2.
In summary, we show for the first time that higher expression of PAX2 in tumors of low malignant potential and low-grade ovarian serous carcinomas than in high-grade carcinomas. Identification of higher PAX2 expression in low malignant potential tumors and low-grade ovarian cancer enhances the current literature hypothesizing differential development pathways between low- and high-grade ovarian carcinomas with low malignant potential tumors potentially developing along a disease continuum to low-grade cancers. Additional studies are being conducted to evaluate the mechanism of PAX2 function in ovarian carcinomas and the potential use of this gene as a therapeutic target that could enable individualized treatment options to be created for patients with ovarian tumors of low malignant potential and low-grade carcinomas.
References
American Cancer Society. Overview: Ovarian Cancer; How many women get ovarian cancer?. 2008 [cited 9-11-2008]; Available from:www.cancer.org.
Seidman JD, Horkayne-Szakaly I, Haiba M, et al. The histologic type and stage distribution of ovarian carcinomas of surface epithelial origin. Int J Gynecol Pathol 2004;23:41–44.
Bertelsen K, Holund B, Andersen E . Reproducibility and prognostic value of histologic type and grade in early epithelial ovarian cancer. Int J Gynecol Cancer 1993;3:72–79.
Malpica A, Deavers MT, Lu K, et al. Grading ovarian serous carcinoma using a two-tier system. Am J Surg Pathol 2004;28:496–504.
Dehari R, Kurman RJ, Logani S, et al. The development of high-grade serous carcinoma from atypical proliferative (borderline) serous tumors and low-grade micropapillary serous carcinoma: a morphologic and molecular genetic analysis. Am J Surg Pathol 2007;31:1007–1012.
Gershenson DM, Sun CC, Lu KH, et al. Clinical behavior of stage II–IV low-grade serous carcinoma of the ovary. Obstet Gynecol 2006;108:361–368.
Auersperg N, Wong AS, Choi KC, et al. Ovarian surface epithelium: biology, endocrinology, and pathology. Endocr Rev 2001;22:255–288.
Levanon K, Crum C, Drapkin R . New insights into the pathogenesis of serous ovarian cancer and its clinical impact. J Clin Oncol 2008;26:5284–5293.
Fathalla MF . Incessant ovulation—a factor in ovarian neoplasia? Lancet 1971;2:163.
Tong GX, Chiriboga L, Hamele-Bena D, et al. Expression of PAX2 in papillary serous carcinoma of the ovary: immunohistochemical evidence of fallopian tube or secondary Mullerian system origin? Mod Pathol 2007;20:856–863.
Piek JM, van Diest PJ, Zweemer RP, et al. Dysplastic changes in prophylactically removed Fallopian tubes of women predisposed to developing ovarian cancer. J Pathol 2001;195:451–456.
Landen Jr CN, Birrer MJ, Sood AK . Early events in the pathogenesis of epithelial ovarian cancer. J Clin Oncol 2008;26:995–1005.
Li C, Wong WH . Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 2001;98:31–36.
Park DC, Yeo SG, Wilson MR, et al. Clusterin interacts with Paclitaxel and confer Paclitaxel resistance in ovarian cancer. Neoplasia 2008;10:964–972.
Silberstein GB, Dressler GR, Van Horn K . Expression of the PAX2 oncogene in human breast cancer and its role in progesterone-dependent mammary growth. Oncogene 2002;21:1009–1016.
Gnarra JR, Dressler GR . Expression of Pax-2 in human renal cell carcinoma and growth inhibition by antisense oligonucleotides. Cancer Res 1995;55:4092–4098.
Singer G, Stohr R, Cope L, et al. Patterns of p53 mutations separate ovarian serous borderline tumors and low- and high-grade carcinomas and provide support for a new model of ovarian carcinogenesis: a mutational analysis with immunohistochemical correlation. Am J Surg Pathol 2005;29:218–224.
O'Neill CJ, Deavers MT, Malpica A, et al. An immunohistochemical comparison between low-grade and high-grade ovarian serous carcinomas: significantly higher expression of p53, MIB1, BCL2, HER-2/neu, and C-KIT in high-grade neoplasms. Am J Surg Pathol 2005;29:1034–1041.
O'Neill CJ, McBride HA, Connolly LE, et al. High-grade ovarian serous carcinoma exhibits significantly higher p16 expression than low-grade serous carcinoma and serous borderline tumour. Histopathology 2007;50:773–779.
Bonome T, Lee JY, Park DC, et al. Expression profiling of serous low malignant potential, low-grade, and high-grade tumors of the ovary. Cancer Res 2005;65:10602–10612.
Wong KK, Lu KH, Malpica A, et al. Significantly greater expression of ER, PR, and ECAD in advanced-stage low-grade ovarian serous carcinoma as revealed by immunohistochemical analysis. Int J Gynecol Pathol 2007;26:404–409.
Longacre TA, McKenney JK, Tazelaar HD, et al. Ovarian serous tumors of low malignant potential (borderline tumors): outcome-based study of 276 patients with long-term (> or =5-year) follow-up. Am J Surg Pathol 2005;29:707–723.
Crispens MA, Bodurka D, Deavers M, et al. Response and survival in patients with progressive or recurrent serous ovarian tumors of low malignant potential. Obstet Gynecol 2002;99:3–10.
Silva EG, Gershenson DM, Malpica A, et al. The recurrence and the overall survival rates of ovarian serous borderline neoplasms with noninvasive implants is time dependent. Am J Surg Pathol 2006;30:1367–1371.
Chi N, Epstein JA . Getting your Pax straight: Pax proteins in development and disease. Trends Genet 2002;18:41–47.
Robson EJ, He SJ, Eccles MR . A PANorama of PAX genes in cancer and development. Nat Rev Cancer 2006;6:52–62.
Tong GX, Melamed J, Mansukhani M, et al. PAX2: a reliable marker for nephrogenic adenoma. Mod Pathol 2006;19:356–363.
Eccles MR, Schimmenti LA . Renal-coloboma syndrome: a multi-system developmental disorder caused by PAX2 mutations. Clin Genet 1999;56:1–9.
Khoubehi B, Kessling AM, Adshead JM, et al. Expression of the developmental and oncogenic PAX2 gene in human prostate cancer. J Urol 2001;165:2115–2120.
Dehbi M, Ghahremani M, Lechner M, et al. The paired-box transcription factor, PAX2, positively modulates expression of the Wilms' tumor suppressor gene (WT1). Oncogene 1996;13:447–453.
Buttiglieri S, Deregibus MC, Bravo S, et al. Role of Pax2 in apoptosis resistance and proinvasive phenotype of Kaposi's sarcoma cells. J Biol Chem 2004;279:4136–4143.
Torban E, Eccles MR, Favor J, et al. PAX2 suppresses apoptosis in renal collecting duct cells. Am J Pathol 2000;157:833–842.
Igarashi T, Ueda T, Suzuki H, et al. Aberrant expression of Pax-2 mRNA in renal cell carcinoma tissue and parenchyma of the affected kidney. Int J Urol 2001;8:60–64.
Muratovska A, Zhou C, He S, et al. Paired-Box genes are frequently expressed in cancer and often required for cancer cell survival. Oncogene 2003;22:7989–7997.
Hueber PA, Waters P, Clark P, et al. PAX2 inactivation enhances cisplatin-induced apoptosis in renal carcinoma cells. Kidney Int 2006;69:1139–1145.
Acknowledgements
We thank Diane Hackett for editing this article and Joseph Celestino for tissue retrieval from tumor tissue bank. This work was supported by grants from the Gynecologic Cancer Foundation, The University of Texas MD Anderson Cancer Center Specialized Program of Research Excellence in Ovarian Cancer Grant P50 CA08369, NIH Research Training Grant T32 CA101642 and NIH R01 CA133057.
Author information
Authors and Affiliations
Corresponding author
Additional information
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on Modern Pathology website (http://www.nature.com/modpathol)
Supplementary information
Rights and permissions
About this article
Cite this article
Tung, C., Mok, S., Tsang, Y. et al. PAX2 expression in low malignant potential ovarian tumors and low-grade ovarian serous carcinomas. Mod Pathol 22, 1243–1250 (2009). https://doi.org/10.1038/modpathol.2009.92
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2009.92
Keywords
This article is cited by
-
PAX2, PAX8, and PR are correlated with ovarian seromucinous borderline tumor with endometriosis
Journal of Ovarian Research (2022)
-
The epigenetic factor BORIS (CTCFL) controls the androgen receptor regulatory network in ovarian cancer
Oncogenesis (2019)
-
PAX8 activates a p53-p21-dependent pro-proliferative effect in high grade serous ovarian carcinoma
Oncogene (2018)
-
Ovarialkarzinom und Präkanzerosen
Der Gynäkologe (2015)
-
Uncovering molecular events associated with the chemosuppressive effects of flaxseed: a microarray analysis of the laying hen model of ovarian cancer
BMC Genomics (2014)