Abstract
Unlike melanoma, clear cell sarcoma harbors either a t(12;22)(q13;q12) recurrent translocation, resulting in an EWSR1/ATF1 chimeric gene, or less commonly a t(2;22)(q34;q12) translocation fusing EWSR1 and CREB1. Few studies have examined the prevalence of all chimeric types and variants to assess the usage of ancillary genetic testing in routine diagnosis. We investigated rearrangement prevalence in 17 clear cell sarcomas, two positive control cell lines, and two melanomas (negative controls). Fluorescence in situ hybridization (FISH) analysis using the LSI EWSR1 break-apart probe and a reverse transcription polymerase chain reaction (RT–PCR) assay optimized for formalin-fixed paraffin-embedded tissue to detect all four reported EWSR1/ATF1 clear cell sarcoma chimeric types and the EWSR1/CREB1 variant was performed. All 15 cases available for testing by FISH were positive for EWSR1 rearrangement including two cases with insufficient RNA for RT–PCR. Thirteen of 15 cases successfully tested by RT–PCR harbored a type 1 chimeric transcript (EWSR1 exon 8/ATF1 exon 4), of which five tumors simultaneously carried a type 2 chimeric transcript (EWSR1 exon 7/ATF1 exon 5). One case carried a type 2 transcript alone and one case contained an EWSR1/CREB1 transcript. Both control cases were positive by both techniques with one case carrying both types 1 and 2 chimeric transcripts and the other types 2 and 3 (EWSR1 exon 10/ATF1 exon 5). Consequently, both techniques are equally effective in assessing for an EWSR1 rearrangement and are useful ancillary diagnostic tests for clear cell sarcoma. They also reinforce the prevalence of this translocation in these tumors. In addition, EWSR1-CREB1 was identified in a clear cell sarcoma of soft tissue providing further evidence that this chimeric variant is not exclusive to gastrointestinal clear cell sarcomas and should be included in RT–PCR assays of soft tissue clear cell sarcomas.
Similar content being viewed by others
Main
Clear cell sarcoma is a rare aggressive sarcoma showing neuroectodermal and melanocytic differentiation. It occurs in young adults typically in the third to fourth decades and preferentially arises in the extremities with the majority being deep seated and involving tendons and aponeuroses.1, 2, 3 Occasionally, primary clear cell sarcoma can arise in visceral organs including the gastrointestinal tract.1, 4, 5, 6, 7, 8, 9, 10 It is one of the few sarcomas with a high propensity for lymph node metastases with frequencies of up to 50% reported.1, 2, 3
Histologically, clear cell sarcoma is characterized by polygonal to spindled cells arranged in nests and fascicles with clear to eosinophillic granular cytoplasm and prominent nucleoli. Scattered wreath-like multinucleate giant cells can also be seen. By immunohistochemical studies, the tumor cells show melanocytic differentiation, being reactive for S-100 protein, HMB45, Melan-A, and microopthalmia transcription factor.1, 2, 3 Interestingly, clear cell sarcomas of the gastrointestinal origin can be negative for specific melanocytic markers.5, 6, 8, 9, 10 In addition, 50% of traditional extremity clear cell sarcomas can harbor pigment.3 This can sometimes make the distinction between clear cell sarcoma and melanoma difficult, especially in situations in which there is no clear in situ component identified or in metastatic melanomas of unknown primary.
Clear cell sarcomas, unlike melanoma, are genetically characterized by a recurrent translocation involving the EWSR1 gene located on chromosome 22q12.11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22 The most common translocation partner is the activating transcription factor-1 (ATF1) gene on chromosome 12q13, of which four chimeric types have been described.11, 12, 13, 15 A second less common translocation partner is CREB1 located on chromosome 2q34, which results in a rearrangement t(2;22) that has been preferentially reported in clear cell sarcomas of the gastrointestinal tract and not in clear cell sarcomas of the soft tissue, until recently.13 Only one EWSR1-CREB1 fusion type has been described so far.10, 13
To date, few studies have examined and characterized all of the EWSR1/ATF1 chimeric types and EWSR1/CREB1 chimeric variant in clear cell sarcomas. The purpose of our study was to examine the usage of both reverse transcription polymerase chain reaction (RT–PCR) and EWSR1 fluorescence in situ hybridization (FISH) on formalin-fixed paraffin-embedded tissue as an ancillary diagnostic tool for clear cell sarcoma and to analyze the chimeric types and variants by RT–PCR.
Materials and methods
With institutional review board approval, 17 clear cell sarcomas from 16 patients (two tumors from the same patient, both metastases from different times to different sites), 2 controls, and 2 melanomas (used as negative controls) with formalin-fixed paraffin-embedded tissue available were retrieved from the pathology files of The University of Texas M.D. Anderson Cancer Center and the Texas Children Hospital/Baylor College of Medicine from 1988 to 2008. Two cases from one patient were published earlier and included in this series.23 Cases were reviewed by a soft tissue pathologist at M.D. Anderson (AJL) and clinical information was obtained when available. Inclusion criteria included tumors composed of spindle to polygonal cells arranged in fascicles or nests, typically involving the subcutis and/or fascia and tendons with immunohistochemical evidence of melanocytic differentiation (S-100 protein and/or HMB45 or Melan-A expression).1, 2, 3, 24 Features of primary cutaneous melanoma such as an associated intraepidermal component or a former history of melanoma resulted in exclusion. All cases were tested in a blinded fashion. Thick sections (4 μm) from representative blocks of tumor (7 metastatic, 9 primary, and 1 of uncertain origin) were prepared on positively charged slides for FISH studies. In addition, two rolls of tumor of 20 μm thick each, or scrapped from multiple 4 μm thick unstained slides, when the tissue block was not available, were used for extraction of total RNA.
To detect and determine the type of EWSR1/ATF1 and EWSR1/CREB1 chimeric transcripts in formalin-fixed archival specimens, an RT–PCR assay was developed. Deparaffination and total RNA extraction was performed using the Ambion Optimum formalin-fixed paraffin-embedded tissue RNA Isolation Kit (Ambion, Austin, TX, USA); 2 μl of total RNA was reverse transcribed into cDNA using a 20-μl reaction volume containing 1.1 μl of random hexamers, 1.1 μl DEPC water, 2.2 μl 5 × RT-buffer, 2.2 μl dNTP (10 mm), 2.2 μl 0.1 M DTT, 0.5 μl Rnase Out (40 μ/ml), and 0.5 μl Super Script Reverse Transcription III. The integrity of RNA was evaluated by running a parallel polymerase chain reaction (PCR) for a 234-bp fragment of the ubiquitously expressed β-actin gene with two primers, one forward (5′-GAGCGGCAAATCGTGCGTGACATT-3′) and one reverse (5′-GATGGAGTTGAAGGTGATTTCGTG-3′). Cases with insufficient RNA quality were excluded from the assay and no PCR was performed. Six different specific PCRs (four reactions for each type of EWSR1-ATF1 chimeric transcript and two reactions for EWSR1-CREB1, one with a forward primer specific for CREB1 and one forward consensus primer to both CREB1 and ATF1) were performed in all samples using both novel (for EWSR1-ATF1) and earlier reported (for EWSR1-CREB1) forward and reverse primers (see Table 1).10 PCR was performed for 34 cycles at 54, 72, and 95°C. Amplicons were visualized on a 2% agarose gel using ethidium bromide staining. Each reaction was optimized for formalin-fixed paraffin-embedded tumor samples, generating amplicons between 130 and 200 bp, although in a few reactions, larger amplicons could also be detected in tumors carrying two chimeric transcripts. In each case, 5 μl of the PCR product or 5 μl of the final amplification product, after being extracted and purified from the gel (using the Qiagen QIAquick Gel Extraction kit, Qiagen, Valencia, CA, USA), were sequenced with the corresponding forward and reverse primer, using the ABI PRISM 3100-Avant Genetic Analyzer. Electropherograms were edited using Chromas Software (Technelysium, Tewantin, QLD, Australia) and the different break points were analyzed by BLAST, using the expected and earlier published different chimeric types. Both positive control cases were positive by FISH and RT–PCR, with one case carrying both types 1 and 2 chimeric transcripts, and the other harboring types 2 and 3 chimeric transcripts.
The FISH for EWSR1 rearrangement was performed using the LSI EWSR1 dual-color, break-apart probe (Abbott Molecular, Des Plaines, IL, USA) according to the manufacturer's recommendations and as described earlier.25 Tissue sections of 4 μm thick were placed onto coated slides, air dried, and baked overnight at 60°C. Slides were deparaffinized in CitriSolv (Fisher, Vernon Hills, IL, USA) three times for 5 min and then immersed in 100% ethanol twice for 1 min. After air drying, slides were treated in Paraffin Pretreatment solution (Paraffin Pretreatment Kit II, Abbott Molecular/Vysis, Des Plaines, IL, USA) for 10 min at 80°C, washed with purified water for 3 min at room temperature, and treated with protease solution for 25 min at 37°C. Slides were subsequently rinsed in purified water for 3 min, air dried, and put in 2 × saline–sodium citrate buffer at 37°C for 30 min, dehydrated in 70, 85, and 100% ethanol, respectively, and allowed to air dry. Next, 10–20 μl of LSI EWSR1 dual-color break-apart probe (Abbott Molecular) was applied to the slides in an approximately 1 cm2 area selected for a pure tumor population (>90% tumor cells), and hybridization was performed at 37°C overnight in a moist chamber. Excess probe was washed away using 2 × saline–sodium citrate buffer/0.3% NP-40 (Fisher) at 73°C for 2 min, and the nuclei were counterstained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI)/Vectashield (Vector Labs, Burlingame, CA). Slides were analyzed using a multi-filtered fluorescence microscope (Olympus BX5, Applied Imaging) and Cytovision Genus 3.7 software (Applied Imaging, San Jose, CA, USA) following standard procedures. A minimum of 200 cells were scored for the presence of rearranged signals by two pathologists in a blinded fashion. A split signal pattern was considered positive for the EWSR1 rearrangement if the distance between the green and the red signals were greater than the diameter of any one signal (Figure 1).
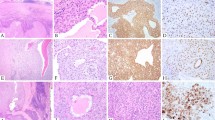
(a–d) Histologic examples of clear cell sarcomas. (a) Cellular neoplasm involving the tendons (20 × ). (b) Tumor composed of epithelioid cells with prominent nucleoli and granular cytoplasm (200 × ). (c) Tumor composed of more spindled cells with multinucleated wreath-like giant cells present (400 × ). (d) Unusual superficial clear cell sarcoma based in the dermis (2 × ). (e and f) Ideogram and FISH with an EWSR1 break-apart probe. Two fluorescently labeled probes (1 (G)reen and 1 (R)ed) flank the EWSR1 gene locus. (e) In cells negative for EWSR1 rearrangement, there are two yellow signals with the green and red signals fused. (f) Cells with 1 green, 1 red signal (ie split signals), and 1 yellow (fusion) signal were considered positive for EWSR1 rearrangement. (g) Sequencing of chimeric transcripts. Type 1 (EWSR1 exon 8/ATF1 exon 4), type 2 (EWSR1 exon 7/ATF1 exon 5), type 3 (EWSR1 exon 10/ATF1 exon 5), and EWSR1-CREB1 were confirmed by sequencing. No type 4 (EWSR1 exon 7/ATF1 exon 7) was identified in our series.
Results
Clinical and Histological Findings
The mean patient age was 29 years (7–66 years) with a male to female ratio of 3:5. The clinical findings are summarized in Table 2. Of the 16 patients, the most common primary site was the foot and lower extremity (8) with other sites including the knee (3), elbow (1), rectum (1), breast (1), palm (1), and forearm (1). All cases were selected to be histologically and clinically consistent with clear cell sarcoma (Figure 1). The majority of the primary tumors were deep seated, with the exception of one tumor, which was centered completely in the dermis with no involvement of the deep soft tissue. This is unusual as the majority of clear cell sarcomas involve the deep soft tissue with some tumors occasionally involving the dermis by direct extension. In 14 of 17 tumors, immunohistochemical studies revealed tumor immunoreactivity for both HMB45 and S-100 protein. Of the three remaining cases, one case had only immunohistochemical study for S-100 protein available and was reactive. The remaining two tumors were also reactive for S-100 protein and negative for HMB45, with one case showing focal reactivity for MART-1.
None of the patients included in the study had a former history of melanoma. Twelve patients had follow-up with an average time of 67 months (6–312 months). Four of the 12 (33%) patients were alive with disease, 5 of 12 (42%) patients died of disease, and 3 of 12 (25%) patients had no evidence of disease. Twelve out of 16 patients (75%) developed metastasis with 8/12 (66%) known to develop lymph node metastases. The average time to metastases was 40 months after initial presentation (0–293 months).
Molecular Findings
Fifteen cases had material available for FISH analysis. All 15 out of 15 clear cell sarcomas showed the presence of an EWSR1 rearrangement by FISH. The results are summarized in Table 2.
Total RNA extraction of 17 clear cell sarcoma specimens yielded RNA of sufficient quality in 15 cases for RT–PCR testing. In two older specimens (>14 years old from 1988 and 1994), only highly degraded RNA insufficient for further testing was retrieved, but these were tested and found to have EWSR1 rearrangements by FISH. The assay detected chimeric transcripts in samples with an RNA concentration as low as 20.61 ng/μl. Thirteen tumors showed a type 1 chimeric transcript (EWSR1 exon 8/ATF1 exon 4), among which five tumors also simultaneously carried a type 2 chimeric transcript (EWSR1 exon 7/ATF1 exon 5). One tumor carried a type 2 chimeric transcript only. One dermally based primary tumor from the palm diagnosed in a 66-year-old patient harbored an EWSR1/CREB1 chimeric transcript. Examples of gel products are shown in Figure 2. In the two different metastases (lung and forearm) from the same patient, excised at different times (15 months apart), one of the tumors harbored both types 1 and 2 chimeric transcripts, whereas the second one carried only a type 1. No type 4 (EWSR1 exon 7, ATF1 exon 7) chimeric transcript was identified in our cases. All amplification products were sequenced for break point characterization. Both melanomas were negative for rearrangements by both molecular tests.
Agarose electrophoresis gels showing RT–PCR products. (a) EWSR1-ATF1. Four separate reactions were performed for each of the four chimeric transcripts: Rx#1 for type 1 (135 bp), Rx#2 for type 2 (132 bp), Rx#3 for type 3 (175 bp). Rx#4 is not shown. Although all specimens were tested for a type 4 chimeric transcript, a type 4 was not encountered in this study. (b) Agarose gel of a tumor harboring two chimeric transcripts. Occasionally, the same reaction simultaneously detected the presence of two chimeric transcripts. Rx#2 reveals the presence of a type 2 (132 bp) and a Type 1 (450 bp). (c) EWSR1-CREB1. Two reactions were run for EWSR1-CREB1. One reaction with a forward primer to an ATF1 and CREB1 consensus sequence (Rx#1, 130 bp) and one with a forward primer specific to CREB1 only (Rx#2, 210 bp). (d) Diagram showing the four EWSR1-ATF1 fusion transcript types and the EWSR-CREB1 fusion variant. Overlying arrows indicate placement of primers. (M, molecular marker; NC, negative control; RC, reagent control; Rx, reaction).
Discussion
Clear cell sarcoma shares many features with melanoma, including histological, immunophenotypic, ultrastructural, and similarities in gene expression patterns.1, 2, 3, 26 However, clear cell sarcoma is genetically distinct lacking melanoma-associated BRAF mutations27 and instead harboring recurrent and characteristic chromosomal translocations involving the EWSR1 gene. In the majority of cases, EWSR1 is fused to ATF1 located on 12q13,11, 12, 13, 15 whereas only rare cases have CREB1 located on 2q34 as the chimeric partner.10, 13 The resulting translocation fuses the 5′ end of the EWSR1 gene to the 3′ end of either ATF1 or, less commonly, CREB1 (Figure 3).
Ideogram showing translocations associated with clear cell sarcoma. Clear cell sarcoma is characterized by recurrent translocations involving EWSR1 located on chromosome 22q12. (a) The most common chimeric partner is ATF1 located on chromosome 12q13. Four EWSR1-ATF1 chimeric transcript types have been described. (b). A less common fusion partner is CREB1 located on chromosome 2q32. Only one ESWR1-CREB1 chimeric transcript type is described.
EWSR1 encodes for an RNA-binding protein and is involved in the recurrent translocations associated with a number of sarcomas including the Ewing family of tumors, desmoplastic small round cell tumor, extraskeletal myxoid chondrosarcoma, angiomatoid fibrous histiocytoma, myxoid liposarcoma (rarely), and clear cell sarcoma.25, 28 ATF1 and CREB1 (cyclic AMP responsive-binding protein) both encode basic leucine zipper transcription factors that are involved in cAMP and Ca+2-induced transcriptional activation.29 As a result, both EWSR1-ATF1 and EWSR1-CREB1 chimeric proteins are believed to likely have a similar function in clear cell sarcoma oncogenesis.10, 30 Interestingly, these translocations are not exclusive to clear cell sarcoma. Angiomatoid fibrous histiocytoma is also characterized by the same recurrent translocations, with EWSR1-CREB1 believed to be most prevalent translocation in these tumors.30 Unlike clear cell sarcoma, angiomatoid fibrous histiocytoma is characterized by sheets of spindled and histiocytoid cells associated with hemorrhage and usually a prominent peripheral lymphocyte infiltrate, which are reactive to various degrees for desmin, CD68, CD99, and epithelial membrane antigen. These tumors do not have melanocytic differentiation and have an overall excellent prognosis.3, 31, 32 The discrepant phenotype and clinical behavior between these two neoplasms suggests that, even though these chimeric genes may have a function in early tumor development of both, other factors such as a different cell of origin and/or additional oncogenic genetic events may be associated and perhaps required for the development of a full tumor phenotype on each of these tumors.
Owing to the characteristic and recurrent association of these translocations and clear cell sarcoma, molecular diagnostics can serve as a useful diagnostic ancillary tool, especially in differentiating melanoma from clear cell sarcoma. Using FISH, Patel et al.14 found that 7/10 (70%) clear cell sarcomas harbored a rearrangement in the EWSR1 locus, whereas none of the 32-tested melanomas carried the rearrangement. In contrast, all 15 clear cell sarcomas in our series, with material available, were positive for EWSR1 rearrangement by FISH. The discrepancy in the results may emphasize the difficulty in differentiating metastatic melanoma of unknown primary from clear cell sarcoma.
RT–PCR has also been found to be valuable not only detecting, but also typing clear cell sarcoma chimeric transcripts. Chimeric transcript detection rate in clear cell sarcoma by this method varies from 93 to 100%.11, 12, 13, 15 (Table 3) Minor discrepancies in detection rates may be partially explained by the fact that not all studies included the less prevalent chimeric transcripts in their RT–PCR assays. Coindre et al.12 found chimeric transcripts in 38 out of 41 (93%) formalin-fixed paraffin-embedded samples tested using a real-time RT–PCR assay for types 1 and 2 EWSR1-ATF1 chimeric transcripts. None of 14 tested melanomas in their study showed either transcript. Using RT–PCR to detect types 1–3 EWSR1-ATF1 chimeric transcripts, Antonescu et al.15 found chimeric transcripts in 11/12 (92%) frozen or paraffin-embedded tumors, whereas Panagopoulos et al.11 found all 10 paraffin-embedded formalin-fixed tumors contained at least one of the four EWSR1-ATF1 chimeric transcripts. Aside from this study, only Hisaoka et al.13 have examined for the presence of all four chimeric EWSR1-ATF1 as well as EWSR1-CREB1 chimeric transcripts using an RT–PCR assay. In their study, all 31 cases were positive. In our series, all cases with sufficient RNA (15/15) were positive for a known clear cell sarcoma chimeric transcript. The two cases with insufficient RNA were both from samples older than 14 years of age, in which the RNA was most likely degraded. The oldest successfully tested sample in our series was 10 years. In both cases in which RT–PCR failed to detect a chimeric transcript, an EWSR1 rearrangement was detected by FISH, highlighting the usefulness of FISH for the diagnosis of specimens with inadequate or degraded RNA.
Having access to multiple methods for detecting fusion genes or transcripts can be helpful as both FISH and RT–PCR are susceptible to failure or false negative in certain situations. In general, RT–PCR is more sensitive to nucleic acid integrity. The age of the sample, the use of decalcification, and delays in fixation or over fixation can all affect the ability to retrieve adequate RNA for diagnosis. Stringent laboratory protocols must be also observed in RT–PCR to prevent cross-contamination between samples because of its sensitivity. In contrast, FISH is more robust technique, but harsh treatment of the tissue (extensive decalcification for instance) can also affect its performance. Unlike RT–PCR, it can provide for in situ confirmation directly on tissue sections and many FISH probes, including the EWSR1 break-apart probe used in this study, are commercially available. It also does not require knowledge of the fusion partner, which is useful if an RT–PCR assay does not cover the more rare fusion partners. Conversely, break-apart FISH does not provide information about the fusion partner, which can be diagnostically relevant in certain sarcomas. As discussed above, different sarcomas may carry rearrangements of the same locus and thus break-apart FISH is not entirely specific, a fact that is particularly relevant in limited biopsies.
On account of the variability of EWSR1 and ATF1 intronic break points, four EWSR1-ATF1 chimeric variants have been described.11, 13 Type 1 resulting from the fusion of EWSR1 exon 8 and ATF1 exon 4 is the most common chimeric transcript in most studies, with the second most common being type 2 (EWSR1 exon 7 and ATF1 exon 5). (Table 3.) The remaining EWSR1-ATF1 chimeric transcript types, types 3 (EWSR1 exon 10 and ATF1 exon 5) and 4 (EWSR1 exon 7 and ATF1 exon 7) are exceedingly rare with type 4 found in only a single case, which also harbored multiple chimeric transcripts.11, 12, 13, 15 In addition, multiple chimeric transcripts in the same tumor have been described in other studies,11, 12, 13 suggesting two clones or perhaps more likely, post-transcriptional alternative splicing. In our series, type 1 was the most common chimeric transcript identified alone (8/15, 53%). Only a single case harbored a type 2 (1/15, 7%) alone. Similar to other studies, tumors with multiple chimeric transcripts were identified (5/15, 33%) in our series, all of which were a combination of types 1 and 2. Two different metastases in the same patient diagnosed at separate times and from different sites also showed differences in chimeric transcript with one case showing type 1 alone and the other showing both types 1 and 2 (cases 11 and 12). This finding may represent the presence of different metastatic tumor clones. Alternatively, it may indicate that there are differences in the transcription or post-transcription modification efficiency between the various chimeric transcript types resulting in low level transcript levels (below the level of detection) in one of the earlier seen chimeric transcript types. No type 3 or type 4 chimeric transcripts was identified in our series, except for one control case, which harbored a type 3 chimeric transcript in combination with a type 1. Interestingly, Coindre et al.12 found more type 2 transcripts (32/38, 84% of 12 with type 2 alone and 20 in combination with type 1) than type 1 (24/38, 64% of only 4 with type 1 alone and 20 in combination with type 2.) The discrepancy between this study and the others is uncertain, but perhaps represents variation because of sample size. So far, there is no prognostic association with chimeric transcript(s) type in clear cell sarcoma.12, 13 Our series lack sufficient sample size and prognostic correlation and virtually all of our tumors behaved aggressively, as reported earlier.1, 2, 3, 24, 33, 34
Overall, the role of chimeric transcript types and variants in sarcomas is uncertain. One example is synovial sarcoma, in which SYT-SSX chimeric transcript type I was found to be significantly associated with a worse prognosis in some studies, but not in others.35, 36 However, there is data that chimeric transcript type may play a role in targeted therapy in some sarcomas. Preliminary studies have suggested that myxoid liposarcomas with type I FUS-CHOP chimeric transcript may respond better to trabectedin.37, 38 Although the role of chimeric transcript type and variants have not been established in clear cell sarcoma, future studies or new therapeutic approaches may be revealing.
Unlike EWSR1-ATF1, only one chimeric variant for EWSR1-CREB1 has been reported. Originally, this chimeric transcript variant was suggested to be preferential to clear cell sarcoma arising in the gastrointestinal tract based on the observation of three cases in this rare type of clear cell sarcoma.10 However, two non-gastrointestinal clear cell sarcomas with EWSR1-CREB1 have been recently described by Hisako et al.13 In our series, we report the third case of a clear cell sarcoma of soft tissue, which was not only associated with an EWSR1-CREB1 chimeric transcript, but also unusual in that it was dermally based on the palm. Consequently, EWSR1-CREB1 is a rare chimeric transcript, which can be found in all clear cell sarcomas regardless of site and can be missed if only RT–PCR for EWSR1-ATF1 chimeric transcripts is performed. FISH would still be positive for a rearrangement in EWSR1, as in our case, but is not specific for the chimeric partner. Both EWSR1-ATF1 and EWSR1-CREB1 are known to occur in gastrointestinal clear cell sarcomas.5, 6, 8, 10 However, the prevalence of chimeric transcript types in gastrointestinal clear cell sarcomas at this site remains uncertain given the rare nature of this tumor.
Our study shows that both an RT–PCR assay and a break-apart FISH for EWSR1 are useful molecular diagnostic tests for clear cell sarcoma. RT–PCR is useful for the identification of EWSR1-specific partner gene, break point, and chimeric transcript types. FISH is especially useful in older specimens, in which RNA is often degraded. Similar to other studies, EWSR1-ATF1 chimeric transcripts, specifically type 1, was the most common chimeric transcript found, with many cases carrying multiple chimeric transcripts. Furthermore, we present further evidence that EWSR1-CREB1 is not exclusive to gastrointestinal tract tumors, indicating the need of incorporating this variant in routine molecular testing of soft tissue clear cell sarcoma specimens if RT–PCR for EWSR1-ATF1 is negative. Further studies are needed to determine the biological meaning and potential prognostic significance of the different chimeric types.
References
Weiss SW, Goldblum JR . Enzinger & Weiss's Soft Tissue Tumor. 5th edn. Mosby Elsevier: St Louis, 2008, pp. 926–934.
Meis-Kindblom JM . Clear cell sarcoma of tendons and aponeuroses: a historical perspective and tribute to the man behind the entity. Adv Anat Pathol 2006;13:286–292.
Fletcher CDM, Unni KK, Mertens F, (eds). World Health Organization of Tumors. Pathology and Genetics of Tumours of Soft Tissue and Bone, 2002 edn. IARC Press: Lyon, France, 2002, pp. 211–212.
Abdulkader I, Cameselle-Teijeiro J, de Alava E, et al. Intestinal clear cell sarcoma with melanocytic differentiation and EWS [corrected] rearrangement: report of a case. Int J Surg Pathol 2008;16:189–193.
Comin CE, Novelli L, Tornaboni D, et al. Clear cell sarcoma of the ileum: report of a case and review of literature. Virchows Arch 2007;451:839–845.
Lyle PL, Amato CM, Fitzpatrick JE, et al. Gastrointestinal melanoma or clear cell sarcoma? Molecular evaluation of 7 cases previously diagnosed as malignant melanoma. Am J Surg Pathol 2008;32:858–866.
Marcon N, Montagne K, Corby S, et al. Primary clear cell sarcoma of the ileum. Ann Pathol 2007;27:369–372.
Taminelli L, Zaman K, Gengler C, et al. Primary clear cell sarcoma of the ileum: an uncommon and misleading site. Virchows Arch 2005;447:772–777.
Venkataraman G, Quinn AM, Williams J, et al. Clear cell sarcoma of the small bowel: a potential pitfall. Case report. APMIS 2005;113:716–719.
Antonescu CR, Nafa K, Segal NH, et al. EWS-CREB1: a recurrent variant fusion in clear cell sarcoma—association with gastrointestinal location and absence of melanocytic differentiation. Clin Cancer Res 2006;12:5356–5362.
Panagopoulos I, Mertens F, Debiec-Rychter M, et al. Molecular genetic characterization of the EWS/ATF1 fusion gene in clear cell sarcoma of tendons and aponeuroses. Int J Cancer 2002;99:560–567.
Coindre JM, Hostein I, Terrier P, et al. Diagnosis of clear cell sarcoma by real-time reverse transcriptase-polymerase chain reaction analysis of paraffin embedded tissues: clinicopathologic and molecular analysis of 44 patients from the French sarcoma group. Cancer 2006;107:1055–1064.
Hisaoka M, Ishida T, Kuo TT, et al. Clear cell sarcoma of soft tissue: a clinicopathologic, immunohistochemical, and molecular analysis of 33 cases. Am J Surg Pathol 2008;32:452–460.
Patel RM, Downs-Kelly E, Weiss SW, et al. Dual-color, break-apart fluorescence in situ hybridization for EWS gene rearrangement distinguishes clear cell sarcoma of soft tissue from malignant melanoma. Mod Pathol 2005;18:1585–1590.
Antonescu CR, Tschernyavsky SJ, Woodruff JM, et al. Molecular diagnosis of clear cell sarcoma: detection of EWS-ATF1 and MITF-M transcripts and histopathological and ultrastructural analysis of 12 cases. J Mol Diagn 2002;4:44–52.
Bridge JA, Sreekantaiah C, Neff JR, et al. Cytogenetic findings in clear cell sarcoma of tendons and aponeuroses. Malignant melanoma of soft parts. Cancer Genet Cytogenet 1991;52:101–106.
Fletcher JA . Translocation (12;22)(q13-14;q12) is a nonrandom aberration in soft-tissue clear-cell sarcoma. Genes Chromosomes Cancer 1992;5:184.
Reeves BR, Fletcher CD, Gusterson BA . Translocation t(12;22)(q13;q13) is a nonrandom rearrangement in clear cell sarcoma. Cancer Genet Cytogenet 1992;64:101–103.
Rodriguez E, Sreekantaiah C, Reuter VE, et al. t(12;22)(q13;q13) and trisomy 8 are nonrandom aberrations in clear-cell sarcoma. Cancer Genet Cytogenet 1992;64:107–110.
Stenman G, Kindblom LG, Angervall L . Reciprocal translocation t(12;22)(q13;q13) in clear-cell sarcoma of tendons and aponeuroses. Genes Chromosomes Cancer 1992;4:122–127.
Pellin A, Monteagudo C, Lopez-Gines C, et al. New type of chimeric fusion product between the EWS and ATF1 genes in clear cell sarcoma (malignant melanoma of soft parts). Genes Chromosomes Cancer 1998;23:358–360.
Speleman F, Delattre O, Peter M, et al. Malignant melanoma of the soft parts (clear-cell sarcoma): confirmation of EWS and ATF-1 gene fusion caused by a t(12;22) translocation. Mod Pathol 1997;10:496–499.
Curry CV, Dishop MK, Hicks MJ, et al. Clear cell sarcoma of soft tissue: diagnostic utility of fluorescence in situ hybridization and reverse transcriptase polymerase chain reaction. J Cutan Pathol 2008;35:411–417.
Enzinger FM . Clear-cell sarcoma of tendons and aponeuroses. an analysis of 21 cases. Cancer 1965;18:1163–1174.
Wang WL, Mayordomo E, Czerniak BA, et al. Fluorescence in situ hybridization is a useful ancillary diagnostic tool for extraskeletal myxoid chondrosarcoma. Mod Pathol 2008;21:1303–1310.
Segal NH, Pavlidis P, Noble WS, et al. Classification of clear-cell sarcoma as a subtype of melanoma by genomic profiling. J Clin Oncol 2003;21:1775–1781.
Panagopoulos I, Mertens F, Isaksson M, et al. Absence of mutations of the BRAF gene in malignant melanoma of soft parts (clear cell sarcoma of tendons and aponeuroses). Cancer Genet Cytogenet 2005;156:74–76.
Lazar A, Abruzzo LV, Pollock RE, et al. Molecular diagnosis of sarcomas: chromosomal translocations in sarcomas. Arch Pathol Lab Med 2006;130:1199–1207.
Persengiev SP, Green MR . The role of ATF/CREB family members in cell growth, survival and apoptosis. Apoptosis 2003;8:225–228.
Antonescu CR, Dal Cin P, Nafa K, et al. EWSR1-CREB1 is the predominant gene fusion in angiomatoid fibrous histiocytoma. Genes Chromosomes Cancer 2007;46:1051–1060.
Fletcher CD . Angiomatoid ‘malignant fibrous histiocytoma’: an immunohistochemical study indicative of myoid differentiation. Hum Pathol 1991;22:563–568.
Smith ME, Costa MJ, Weiss SW . Evaluation of CD68 and other histiocytic antigens in angiomatoid malignant fibrous histiocytoma. Am J Surg Pathol 1991;15:757–763.
Lucas DR, Nascimento AG, Sim FH . Clear cell sarcoma of soft tissues. Mayo Clinic experience with 35 cases. Am J Surg Pathol 1992;16:1197–1204.
Sara AS, Evans HL, Benjamin RS . Malignant melanoma of soft parts (clear cell sarcoma). A study of 17 cases, with emphasis on prognostic factors. Cancer 1990;65:367–374.
Ladanyi M, Antonescu CR, Leung DH, et al. Impact of SYT-SSX fusion type on the clinical behavior of synovial sarcoma: a multi-institutional retrospective study of 243 patients. Cancer Res 2002;62:135–140.
Guillou L, Benhattar J, Bonichon F, et al. Histologic grade, but not SYT-SSX fusion type, is an important prognostic factor in patients with synovial sarcoma: a multicenter, retrospective analysis. J Clin Oncol 2004;22:4040–4050.
Forni C, Minuzzo M, Virdis E, et al. Trabectedin (ET-743) promotes differentiation in myxoid liposarcoma tumors. Mol Cancer Ther 2009;8:449–457.
Bode-Lesniewska B, Frigerio S, Exner U, et al. Relevance of translocation type in myxoid liposarcoma and identification of a novel EWSR1-DDIT3 fusion. Genes Chromosomes Cancer 2007;46:961–971.
Acknowledgements
The authors express their appreciation to Kim-Anh Vu for her assistance in graphic design. Dr. Alexander Lazar is supported by the University of Texas M.D. Anderson Cancer Center Physician-Scientist Program.
Author information
Authors and Affiliations
Corresponding author
Additional information
Conflict of interest
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Wang, WL., Mayordomo, E., Zhang, W. et al. Detection and characterization of EWSR1/ATF1 and EWSR1/CREB1 chimeric transcripts in clear cell sarcoma (melanoma of soft parts). Mod Pathol 22, 1201–1209 (2009). https://doi.org/10.1038/modpathol.2009.85
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2009.85
Keywords
This article is cited by
-
Phosphorylated ATF1 at Thr184 promotes metastasis and regulates MMP2 expression in gastric cancer
Journal of Translational Medicine (2022)
-
SS18-SSX drives CREB activation in synovial sarcoma
Cellular Oncology (2022)
-
Clear Cell Carcinoma in the Oral Cavity with Three Novel Types of EWSR1-ATF1 Translocation: A Case Report
Head and Neck Pathology (2022)
-
Generation of human embryonic stem cell models to exploit the EWSR1-CREB fusion promiscuity as a common pathway of transformation in human tumors
Oncogene (2021)
-
Clear cell sarcoma-like/malignant gastrointestinal neuroectodermal tumor of the tongue: a clinicopathologic and molecular case report
Virchows Archiv (2021)