Abstract
A subset of lung cancers harbors a small inversion within chromosome 2p, giving rise to a transforming fusion gene, EML4-ALK (echinoderm microtubule-associated protein-like 4 gene and the anaplastic lymphoma kinase gene), which encodes an activated tyrosine kinase. We have earlier examined the presence of EML4-ALK by multiplex reverse transcription-polymerase chain reaction in 363 specimens of lung cancer, identifying 11 adenocarcinoma cases featuring the fusion gene. In this study, we clinicopathologically examined the characteristics of the EML4-ALK-positive cases, including the mutation status of EGFR, KRAS, and TP53, and whether they were of thyroid transcription factor-1 (TTF-1) cell lineage or not. Of 11 patients, 4 (36%) with EML4-ALK-positive lung adenocarcinomas who were below 50 years of age were affected by these diseases, as compared with 12 of 242 patients (5.0%) with EML4-ALK-negative lung adenocarcinomas (P=0.00038). EML4-ALK-positive lung adenocarcinomas were characterized by less-differentiated grade (P=0.0082) and acinar-predominant structure (P<0.0001) in histology. Furthermore, the presence of EML4-ALK appears to be mutually exclusive for EGFR and KRAS mutations (P=0.00018), whereas coexisting with TP53 mutations at a low frequency (1/11=9.1%), and correlating with non- or light smoking (P=0.040), in line with the TTF-1 immunoreactivity. Thus, EML4-ALK-positive tumors may form a distinct entity among lung adenocarcinomas, characterized by young onset, acinar histology, no or rare mutations in EGFR, KRAS, and TP53, and a TTF-1 cell lineage, all in agreement with the prevalence in non- or light smokers.
Similar content being viewed by others
Main
Lung cancer is one of the leading causes of cancer death in both men and women worldwide. Activating mutations within epidermal growth factor receptor (EGFR) have been identified in lung cancers,1, 2 and chemical inhibitors for the kinase activity of EGFR have been found effective in treatment of a subset of lung cancer patients harboring such mutations. Interestingly, the tumors for which EGFR inhibitors are effective develop preferentially in populations of Asian ethnicity and non-smokers, and generally lack KRAS mutations.2, 3, 4 Furthermore, such tumors have low rates of smoking-specific TP53 mutations, such as G/C to T/A transversions.5, 6
Recently, we have found a novel transforming fusion gene joining the echinoderm microtubule-associated protein-like 4 gene (EML4) and the anaplastic lymphoma kinase gene (ALK) in four lung adenocarcinomas and one squamous cell carcinoma.7 The EML4-ALK fusion gene is formed by a small inversion within chromosome 2p. The encoded protein contains the N-terminal part of EML4 and the intracellular catalytic domain of ALK. Replacement of the extracellular and transmembrane domain of ALK with a region of EML4 results in constitutive dimerization of the kinase domain and thereby a consequent increase in its catalytic activity.7
More recently, we have identified novel variants for EML4-ALK fusion gene with cDNA screening and multiplex reverse transcription-polymerase chain reaction (RT-PCR), capturing all possible in-frame fusions of EML4 to exon 20 of ALK. By carrying out cDNA screening, we identified variant 3,8 and using multiplex RT-PCR assays, we identified variants 4 and 59 after the first identification of variants 1 and 2. In variant 3, exon 6 of EML4 is joined to exon 20 of ALK. cDNA from variant 4 ligates exon 14 of EML4 to a position within exon 20 of ALK, whereas another cDNA from a variant 5 tumor connects exon 2 of EML4 to exon 20 of ALK. All the new three isoforms of EML4-ALK have a marked oncogenic activity in vitro as well as in vivo.8, 9 The variant 3 was also identified by Rikova et al10 and another new variant, in which exon 15 of EML4 is jointed to a position within exon 20 of ALK, was identified by Koivunen et al.11
Earlier we conducted the first large scale-study to detect EML4-ALK (3.4%) in lung adenocarcinomas and found five fusion-positive cases (two variant 1 and three variant 2) in 149 adenocarcinoma samples.12 At that point in time, only two variants were recognized, and we investigated their clinicopathological characteristics. However, with development of multiplex RT-PCR for detecting all possible in-frame variants, we captured theoretically all EML4-ALK variants and found 11 EML4-ALK-positive cases among 363 lung cancers.9 In this study, we examined the clinicopathological and genetic features of the 11 tumors, and found EML4-ALK lung cancers to be characterized by a lack of EGFR and KRAS mutations, a low rate for TP53 mutations, a thyroid transcription factor-1 (TTF-1)-positive cell lineage, an acinar histology, and young onset.
Materials and methods
Clinical Samples and Pathological Review
The clinical specimens for this study were 11 lung tumors detected in our earlier study, using multiplex RT-PCR and fluorescent in situ hybridization.9 Briefly, samples were obtained from 363 individuals who underwent surgery at the Cancer Institute Hospital (Tokyo, Japan) between May 1997 and February 2004. The 363 lung cancers comprised 253 adenocarcinomas, 7 adenosquamous carcinomas, 72 squamous cell carcinomas, 7 large-cell carcinomas (including 4 large-cell neuroendocrine carcinomas), 2 pleomorphic carcinomas, and 22 small-cell carcinomas. This project was approved by the Institutional Review Board of the Japanese Foundation for Cancer Research, and informed consent was obtained from each study subject. Histological diagnoses were made based on the World Health Organization (WHO) classification.13 However, with its subdivision of lung adenocarcinomas, more than 80% tumors fell into the mixed subtype category. We therefore additionally used a predominance classification of invasive components, which is mostly based on the WHO classification except for the mixed subtype, such as papillary predominant, acinar predominant, etc. In the predominance classification of invasive components, we diagnose by a component that makes up the predominant portion of invasive lesion even if it is <50%. In addition, we used a differentiation grading that was basically according to the former version of the Japanese Lung Cancer Society criteria,14 as performed earlier.15
Immunohistochemical Analysis
Unstained paraffin-embedded sections were depleted of paraffin with xylene, rehydrated through a graded series of ethanol solutions, and subjected to immunohistochemical staining with a mouse monoclonal antibody (ALK1, 1:20, Dako, Carpinteria, CA, USA). Heat-induced antigen retrieval pretreatment was performed with Target Retrieval Solution pH 9.0 (Dako). Immune complexes were detected with the EnVision+DAB system (Dako) with minor modifications.16 TTF-1 was also immunostained using a mouse monoclonal antibody (clone 8G7G3/1, 1:100, Dako), as described earlier.15 Tumors were considered negative if staining was found in <5% of neoplastic cells, partly positive if present in 5–50%, and positive if in more than 50%. The results of immunostaining with TTF-1 were based on nuclear staining of neoplastic cells.
DNA Extraction and Mutation Analysis of EGFR, KRAS, and TP53
Of 253 patients with adenocarcinomas, both EGFR and KRAS data were available for 68 patients and EGFR data alone for further 12 patients, including all the patients with EML4-ALK-positve cases.12, 17 DNA extraction and mutation analysis of EGFR and KRAS were performed as described earlier.17 Mutation analysis of TP53 was also performed as described earlier.18 Genomic DNAs from fresh tumor samples were prepared and exons 4–8 and 10 of the TP53 gene were analyzed by the PCR – single-strand conformation polymorphism and DNA sequencing. For case #4808, TP53 mutation analysis was performed using DNA extracted from formalin-fixed paraffin-embedded tissue and a method based on direct sequencing, because no fresh sample was available for this study at the time of the current study.
Results
Histologically, the 253 adenocarcinomas comprised 213 mixed subtypes, 19 acinar, 9 papillary, 4 solid, 1 other, and 7 bronchioloalveolar carcinomas based on the WHO classification. According to the subdivision of lung adenocarcinomas with the WHO criteria, more than 80% of tumors fell into the mixed subtype category. However, this contains very varied lesions; for example, a tumor comprising solid and acinar adenocarcinoma elements would be expected to have a very different prognosis from one composed of bronchioloalveolar carcinoma, with only a small amount of papillary adenocarcinoma. We therefore additionally used a predominance classification and also paid attention to the minor components. According to the predominance subtypes in adenocarcinomas, 6 of 11 EML4-ALK-positive lung cancers (54.5%) were subclassified as acinar adenocarcinomas (P=0.000044, Table 1), as compared with 4 based on the WHO classification (36.4%, P=0.0018, Table 2). In other words, 6 of 34 (18%) acinar-predominant adenocarcinomas, as well as 4 of 19 (21%) acinar adenocarcinomas based on the WHO classification, have EML4-ALK fusion. In adenocarcinomas not subclassified as acinar adenocarcinomas based on the WHO criteria, acinar structures were also frequently observed (Figure 1). In differentiation grading, EML4-ALK lung cancers were less differentiated (Table 3, P=0.0082, 10/11). In addition, they often featured mucin production, as proven by Alcian Blue staining (Figure 1b) with acinar structures. As for the cell types originally proposed by Hashimoto et al,18 the columnar cell type was characteristic of EML4-ALK lung cancers (Figure 1).
EML4-ALK-positive lung adenocarcinomas were also found to be significantly smaller than other lung adenocarcinomas (Table 3, P=0.031), in line with the lack of bronchioloalveolar components.
Patients with EML4-ALK lung cancers tended to be young (56 vs 64 years for other tumor types, P=0.0062). We defined early-onset lung cancers by classifying patients as below or above 50 years of age. In 253 patients with lung adenocarcinomas, 16 patients were affected by the disease at below 50 years of age. Four of 11 patients (36%) with EML4-ALK-positive lung cancers were affected by these diseases at below 50 years of age, as compared with 12 of 242 patients (5.0%) with EML4-ALK-negative lung cancers (P=0.00038).
It is true that the EML4-ALK translocation was first found in a smoker's lung cancer, but overall there was no significant difference between smokers' and non-smokers' tumors with regard to EML4-ALK fusion (P=0.37). Smoking habits can be classified into the following two grades of cumulative smoking based on the smoking index (SI), a product of the number of cigarettes per day, and the duration in years: (a) non-smokers and light smokers (SI <400); and (b) heavy smokers (SI=400 or above). Ten of the 11 (91%) EML4-ALK-positive lung cancer patients had SI <400, as compared with 109 of 241 (45%) EML4-ALK-negative lung cancer patients (P=0.040). In this study, EML4-ALK fusion was detected in only one heavy smoker's lung cancer (1/11=9.1%).
EGFR and KRAS mutations are mutually exclusive in usual cases while being two major oncogenic drivers of lung adenocarcinoma development. EML4-ALK-positive lung cancers lacked both EGFR and KRAS mutations (P=0.00018), and only 1 of 11 (9.1%) harbored a TP53 mutation (Table 2). It is noteworthy that the single mutation was a G/A transition (GTG → ATG) (V → M) in codon 273, exon 8. This is known to be a spontaneous rather than a tobacco-carcinogen-induced mutation, usually seen in non-smokers' lung cancers.
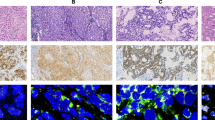
In the EML4-ALK-positive 11 cases, immunohistochemical assays with the anti-ALK antibody ALK1 consistently showed definite staining. As illustrated in Figure 2a, the cytoplasm of tumor cells harboring the variant 2 (tumor ID #2374) was strongly stained with fine granular accentuation. Although we performed the immunostaining of 88 EML4-ALK-negative lung adenocarcinoma specimens, we could discriminate all the fusion-negative specimens from the fusion-positive ones by our refined immunohistochemical condition.16 All the 11 cases were also positive (six cases) or partly positive (five cases) for TTF-1 immunohistochemistry (Figure 2b), a characteristic of alveolar type II cells, which is featured in non-smokers' cancers.
(a) Immunohistochemical analysis with a monoclonal anti-ALK antibody of lung adenocarcinoma specimens with (tumor ID #2374) and without (tumor ID #4227) EML4-ALK fusion. Note the diffuse staining in the cytoplasm with fine granular accentuation apparent for the EML4-ALK-positive tumor. (b) Immunohistochemical analysis of lung adenocarcinoma specimens with EML4-ALK fusion using a monoclonal anti-TTF-1 antibody. The EML4-ALK-positive tumors are partly (ID #2075) or diffusely (ID #7969) positive.
Discussion
With the present large-scale screen for EML4-ALK fusion in lung cancers, we detected 11 adenocarcinomas with an EML4-ALK translocation. In the current study, we revealed a relatively young occurrence and a typically less-differentiated acinar histology, which might be used as clinical pointers. It is of great interest that EML4-ALK translocation is associated with young onset, whereas EGFR mutation status is not associated with the patient's age at diagnosis.4
Currently, anaplastic large-cell lymphomas (ALCLs) are divided into three entities, namely primary systemic ALK (+) ALCL, primary systemic ALK (−) ALCL, and primary cutaneous ALCL. The ALK expression is caused most commonly t(2;5) by chromosomal translocations, and ALK (+) ALCL predominantly affects young male patients and, if treated with chemotherapy, has a favorable prognosis.19 This might similarly be applicable to EML4-ALK lung cancers. Presently, the primary treatment for lung cancers is surgery where possible. However, for EML4-ALK lung cancers, chemotherapy or a targeted therapy with an ALK inhibitor might be effective, given that EML4-ALK-dependent cells are known to undergo apoptosis in response.7, 8, 9, 11
Here, EML4-ALK fusion was found to be mutually exclusive for EGFR or KRAS mutations, thus pointing to a distinct genetic subtype of lung adenocarcinoma. The possibility of a genetic classification of lung adenocarcinomas based on oncogene mutations has already been considered. In fact, one-third to nearly half of Japanese adenocarcinomas harbor EGFR mutations,4, 20 about 10% have KRAS mutations21, 22, 23 and about 4% have EML4-ALK translocations, implying that two-thirds of adenocarcinomas feature mutually exclusive oncogenic mutations. The mutation rate of TP53 (1/11=9.1%) was also low compared with that of lung adenocarcinomas in general (41%),18 and the single mutation found was G to A transition, which was not related to smoking. Strong in vitro as well as in vivo oncogenic activity of EML4-ALK fusion products8, 9 might account for the lack of other genetic alterations.
All 11 EML4-ALK lung cancers were positive or partly positive for TTF-1 immunostaining. TTF-1 has a decisive role as a master regulatory transcription factor in lung development and in maintenance of the functions of terminal respiratory unit (TRU) cells.24 The TTF-1 positivity of EML4-ALK lung cancers suggests that this subtype might have a TRU histogenesis. TRU-type lung cancers with a TTF-1-positive cell lineage often occur in non- or light smokers, which frequently harbor EGFR mutations (61%) and have less-frequent TP53 mutations (36%) as compared with non-TRU-types (57%).22 EML4-ALK lung cancers also occur in non- or light smokers but do not harbor EGFR mutations. The low frequency of TP53 mutations (9.1%) not only indicates strong oncogenic activity for EML4-ALK fusion products but also suggests an independence from smoking, because smoker's adenocarcinomas very frequently harbor TP53 mutations.18
Histologically, less-differentiated acinar structures composed of columnar cells appear characteristic of EML4-ALK lung adenocarcinomas. Generally, the columnar cell type is also found in smoker's lung adenocarcinomas, whereas the hobnail cell type, characterized by cytoplasmic protrusions and with a tadpole shape, is often observed in non-smoker's lung adenocarcinomas.18 Although EML4-ALK lung cancers are TTF-1-positive, their histology is similar to lung cancers developing in smokers, which is interesting in the view of histology–etiology relationships.
Presently, lung adenocarcinomas may be genetically divided into EGFR-mutated, KRAS-mutated, and EML4-ALK-related subtypes. We here elucidated the clinicopathologic, histologic, and genetic characteristics of EML4-ALK lung cancers, bearing etiologic implications in mind. Just as some EGFR-mutated lung cancers can be successfully treated with EGFR inhibitors, EML4-ALK lung cancers may respond to a specific inhibitor treatment, allowing a good prognosis.
References
Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129–2139.
Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497–1500.
Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from ‘never smokers’ and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA 2004;101:13306–13311.
Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst 2005;97:339–346.
Pfeifer GP, Denissenko MF, Olivier M, et al. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene 2002;21:7435–7451.
Suzuki H, Takahashi T, Kuroishi T, et al. p53 mutations in non-small cell lung cancer in Japan: association between mutations and smoking. Cancer Res 1992;52:734–736.
Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561–566.
Choi YL, Takeuchi K, Soda M, et al. Identification of novel isoforms of the EML4-ALK transforming gene in non-small cell lung cancer. Cancer Res 2008;68:4971–4976.
Takeuchi K, Choi YL, Soda M, et al. Multiplex reverse transcription-PCR screening for EML4-ALK fusion transcripts. Clin Cancer Res 2008;14:6618–6624.
Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007;131:1190–1203.
Koivunen JP, Mermel C, Zejnullahu K, et al. EML4–ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res 2008;14:4275–4283.
Inamura K, Takeuchi K, Togashi Y, et al. EML4-ALK fusion is linked to histological characteristics in a subset of lung cancers. J Thorac Oncol 2008;3:13–17.
Travis WD, Brambilla E, Muller-Hermelink HK, et al. World Health Organization Classification of Tumours: Pathology and Genetics of Tumours of the Lung, Pleural, Thymus and Heart. Springer: Berlin, 2004.
Japan Lung Cancer Society. General Rules for Clinical and Pathologic Record of Lung Cancer [in Japanese], 5th edn. Kanahara: Tokyo, 1999.
Inamura K, Satoh Y, Okumura S, et al. Pulmonary adenocarcinomas with enteric differentiation: histologic and immunohistochemical characteristics compared with metastatic colorectal cancers and usual pulmonary adenocarcinomas. Am J Surg Pathol 2005;29:660–665.
Takeuchi K, Choi YL, Togashi Y, et al. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res 2009 (in press).
Inamura K, Togashi Y, Nomura K, et al. Up-regulation of PTEN at the transcriptional level is an adverse prognostic factor in female lung adenocarcinomas. Lung Cancer 2007;57:201–206.
Hashimoto T, Tokuchi Y, Hayashi M, et al. Different subtypes of human lung adenocarcinoma caused by different etiological factors. Evidence from p53 mutational spectra. Am J Pathol 2000;157:2133–2141.
Stein H, Foss HD, Durkop H, et al. CD30(+) anaplastic large cell lymphoma: a review of its histopathologic, genetic, and clinical features. Blood 2000;96:3681–3695.
Kosaka T, Yatabe Y, Endoh H, et al. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res 2004;64:8919–8923.
Noda N, Matsuzoe D, Konno T, et al. K-ras gene mutations in non-small cell lung cancer in Japanese. Oncol Rep 2001;8:889–892.
Yatabe Y, Kosaka T, Takahashi T, et al. EGFR mutation is specific for terminal respiratory unit type adenocarcinoma. Am J Surg Pathol 2005;29:633–639.
Shibata T, Hanada S, Kokubu A, et al. Gene expression profiling of epidermal growth factor receptor/KRAS pathway activation in lung adenocarcinoma. Cancer Sci 2007;98:985–991.
Tanaka H, Yanagisawa K, Shinjo K, et al. Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res 2007;67:6007–6011.
Acknowledgements
The authors thank Dr Malcolm Moore for the English correction of the manuscript, and Ms Kazuko Yokokawa, Mr Motoyoshi Iwakoshi, Ms Miyuki Kogure, and Ms Tomoyo Kakita for their technical assistance, and Ms Yuki Takano for her secretarial work. Parts of this study were supported financially by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, from the Japan Society for the Promotion of Science, and by grants from the Ministry of Health, Labour and Welfare, the Smoking Research Foundation, the National Institute of Biomedical Innovation, and the Vehicle Racing Commemorative Foundation, as well as a Grant-in-Aid for Young Scientists (B).
Author information
Authors and Affiliations
Corresponding author
Additional information
Conflict of interest
K Takeuchi is a consultant providing advisory services to Dako for their antibodies.
Rights and permissions
About this article
Cite this article
Inamura, K., Takeuchi, K., Togashi, Y. et al. EML4-ALK lung cancers are characterized by rare other mutations, a TTF-1 cell lineage, an acinar histology, and young onset. Mod Pathol 22, 508–515 (2009). https://doi.org/10.1038/modpathol.2009.2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2009.2
Keywords
This article is cited by
-
Cancer organoid-based diagnosis reactivity prediction (CODRP) index-based anticancer drug sensitivity test in ALK-rearrangement positive non-small cell lung cancer (NSCLC)
Journal of Experimental & Clinical Cancer Research (2023)
-
Genetic fusions favor tumorigenesis through degron loss in oncogenes
Nature Communications (2021)
-
Relationship between molecular changes in epidermal growth factor receptor (EGFR) and anaplastic lymphoma kinase (ALK) mutations in lung adenocarcinoma*
Oncology and Translational Medicine (2021)
-
Individualized nomogram for predicting ALK rearrangement status in lung adenocarcinoma patients
European Radiology (2021)
-
ALK rearrangement: a high-frequency alteration in ovarian metastasis from lung adenocarcinoma
Diagnostic Pathology (2019)