Abstract
Well-differentiated liposarcoma/atypical lipomatous tumor and dedifferentiated liposarcoma can be difficult to distinguish from benign lipomatous neoplasms and other high-grade sarcomas, respectively. Cytogenetics in these tumors has identified ring and giant chromosomes composed of 12q13-15 amplicons including the MDM2 gene. Identifying MDM2 amplification by fluorescence in situ hybridization may prove an adjunctive tool in the diagnosis of lipomatous neoplasms. Dual color fluorescence in situ hybridization employing a laboratory-developed BAC label probe cocktail specific for MDM2 (12q15) and a probe for the centromeric region of chromosome 12 (Abbott Molecular, DesPlaines, IL) was performed on formalin-fixed and paraffin-embedded tissue including whole sections from atypical lipomatous tumors (n=13), dedifferentiated liposarcomas (n=14), benign lipomatous tumors (n=30), and pleomorphic sarcoma, not otherwise specified (n=10), and a tissue microarray containing a variety of high-grade sarcomas (n=63). An MDM2/chromosome 12 ratio ≥2.0 was considered amplified, <2.0 nonamplified, and cases displaying >2 signals of both probes and an MDM2 ratio <2.0 polysomic for chromosome 12. Of the well-differentiated and dedifferentiated liposarcomas, 100% showed amplification of MDM2. Chromosome 12 polysomy was noted in 89% of spindle cell/pleomorphic lipomas, while all angiolipomas and lipomas were nonamplified and eusomic. MDM2 amplification was observed in 40% of pleomorphic sarcomas and a small subset of high-grade sarcomas (3/63). MDM2/chromosome 12 fluorescence in situ hybridization is a sensitive and specific tool (both 100%) in evaluating low-grade lipomatous neoplasms. The specificity decreases in high-grade sarcomas, as MDM2 amplification was observed in a small portion of pleomorphic sarcomas and high-grade sarcomas other than dedifferentiated liposarcomas. Importantly, none of the benign lipomatous lesions were MDM2 amplified and even cells in areas of well-differentiated liposarcomas with minimal cytologic atypia were amplified, making the probe a valuable tool in the diagnosis of even limited biopsy samples of well-differentiated lipomatous neoplasms.
Similar content being viewed by others
Main
Well-differentiated liposarcoma/atypical lipomatous tumor and dedifferentiated liposarcoma are among the most common malignant soft tissue tumors presenting in older adults.1, 2, 3 These tumors typically arise in the deep soft tissue of the proximal lower extremities as slow growing masses or in the retroperitoneum with symptoms related to the presence of an abdominal mass.4, 5 Well-differentiated liposarcomas/atypical lipomatous tumors are often difficult to distinguish morphologically from benign lipomatous tumors, while dedifferentiated liposarcomas may be challenging to distinguish from other high-grade sarcomas, especially on needle biopsy specimens that lack areas of well-differentiated liposarcomas.
Well-differentiated liposarcoma/atypical lipomatous tumors and dedifferentiated liposarcomas have been shown by cytogenetics to harbor ring and giant marker chromosomes consisting of amplicons of the 12q13-15 region, resulting in amplification of several genes, including most notably MDM2. PCR, comparative genomic hybridization (CGH), and fluorescence in situ hybridization (FISH) can also be used to demonstrate amplification in this chromosomal region.6, 7, 8, 9, 10 MDM2 (murine double minute) gene is an oncogene whose expression plays important roles in controlling the cell cycle and tumorigenesis by degradation of TP53.11 With the increased use of small needle core biopsy and fine needle aspiration (FNA) in which only limited tissue is available for histologic evaluation, increased utilization of molecular studies to identify the characteristic molecular aberrations of well-differentiated liposarcoma/atypical lipomatous tumors and dedifferentiated liposarcomas may prove useful in clinical practice.
A dual-color MDM2 FISH probe was utilized, which permits recognition of MDM2 gene (12q13) amplification in formalin-fixed, paraffin-embedded tissues (FFPET). This probe, coupled with a centromeric reference probe for chromosome 12 (CEP12), could provide an important ancillary diagnostic tool in cases where fresh tissue is either not available for conventional cytogenetic analysis and/or PCR-based studies, or where performing such studies is impractical. Here, we report our experience with this novel probe set to distinguish well-differentiated liposarcoma/atypical lipomatous tumors and dedifferentiated liposarcomas from their morphologic mimics.
Materials and methods
After receiving Institutional Review Board approval, formalin-fixed and paraffin-embedded resection specimens and excisional biopsies were identified from the archives of the Department of Anatomic Pathology at the Cleveland Clinic. Hematoxylin and eosin-stained sections were independently reviewed by two soft tissue pathologists (JRG and MS). The consensus-verified diagnostic categories included well-differentiated liposarcoma/atypical lipomatous tumors (n=13), low-grade dedifferentiated liposarcomas (n=7), high-grade dedifferentiated liposarcomas (n=7), spindle cell/pleomorphic lipoma (n=9), angiolipoma (n=10), lipoma (n=10), lipoblastoma (n=1), pleomorphic sarcoma, not otherwise specified (n=10), myxoid liposarcoma (n=5), extra-skeletal myxoid chondrosarcoma (n=10), myxofibrosarcoma (n=1), clear cell sarcoma (n=4), low-grade fibromyxoid sarcoma (n=10), solitary fibrous tumor/hemangiopericytoma (n=10), malignant melanoma (n=1), epithelioid sarcoma (n=1), neuroblastoma (n=1), Ewing's sarcoma/primitive neuroectodermal tumor (n=3), desmoplastic small round cell tumor (n=1), alveolar rhabdomyosarcoma (n=3), gastrointestinal stromal tumor (n=4), leiomyosarcoma (n=1), synovial sarcoma (n=4), and malignant peripheral nerve sheath tumor (n=4). Whole tissue sections of the lipomatous tumors were used, as the hypocellularity seen in many of these tumors may preclude representative evaluation in tissue microarray format. The remaining soft tissue neoplasms, except the cases of pleomorphic sarcoma, not otherwise specified, were represented on tissue microarray sections consisting of duplicate 1.5 mm diameter tissue cores, as described previously.12 Whole tissue sections of the pleomorphic sarcoma, not otherwise specified were utilized.
All cases were evaluated with a laboratory-developed BAC probe set. BAC DNAs were purchased from Roswell Park Cancer Institute, Buffalo, NY. BAC DNAs used for this study were RP11-775J10 and RP11-450G15 both containing genomic sequence for the MDM2 gene. The probes were generated by nick translation of the BAC DNAs using a kit purchased from Vysis Inc. In a typical labeling reaction, 1 μg of DNA was nick translated in a 50 μl reaction containing 10 μM spectrum green dUTP, 10 μM dTTP, and 20 μM dNTP mix (dATP, dCTP, and dGTP) in 1X nick translation buffer provided with 5 μl of enzyme in 8 h reaction at 15°C. Enzyme was inactivated at 70°C for 10 min and reactions precipitated along with human placental DNA (Sigma). Precipitates were centrifuged in cold for 30 min and resuspended in 20 μl of water. Probes were validated against controls such as normal human tonsil and known MDM2 amplified samples before staining the cohort of sample slides.
The FISH assay was performed according to established laboratory protocol, as described previously.13 Blinded to the histologic diagnosis and using an Olympus IX-50 microscope (Olympus, Tokyo, Japan), cases were scored by counting a minimum of 40 nuclei per case under oil immersion at × 100 magnification with a DAPI/green/red triple band pass filter. Only nuclei with at least two CEP12 signals were evaluated to minimize nuclear truncation artifact that occurs in paraffin-embedded 4-μm sections. Overlapping tumor nuclei were also excluded from evaluation to decrease false-positive scoring. The average number of MDM2 and CEP12 signals was then determined and a MDM2/CEP12 ratio was calculated for each case. A ratio ≥2.0 was considered amplified for the MDM2 gene, while a ratio <2.0 was considered nonamplified. A ratio of <2.0 with >2 signals of both probes was considered polysomic for CEP12.
Results
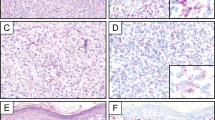
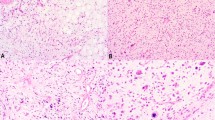
All cases analyzed by FISH had detectable and evaluable nuclear signals. The data presented in Table 1 depict the results for each tumor category. Of the well-differentiated liposarcomas/atypical lipomatous tumors studied, 100% (13/13) showed amplification of MDM2 (mean: >16 signal gains/nucleus; MDM2/CEP12 ratio: 7.2). A total of 14 of 14 (100%) dedifferentiated liposarcomas showed amplification of MDM2 (mean: >18 signal gains/nucleus; MDM2/CEP12 ratio: 8.2). Both the well-differentiated and dedifferentiated areas showed a similar degree of MDM2 amplification (Figure 1). In well-differentiated liposarcomas/atypical lipomatous tumors, MDM2 amplification was identified in both the cytologically atypical and nonatypical cells (Figure 2). CEP12 polysomy was noted in 89% (8/9) spindle cell/pleomorphic lipomas (MDM2/CEP12 ratio: 0.97) (Figure 3). All angiolipomas, lipomas, and the lipoblastoma were not amplified (MDM2/CEP12 ratio: 1) (Figure 3).
(a) Well-differentiated liposarcoma/atypical lipomatous tumor, adipocytes with cytologic atypia cells. (b) MDM2 gene amplification in a well-differentiated liposarcoma/atypical lipomatous tumor (many MDM2-red signals and only three CEP12-green signals). (c) Dedifferentiated liposarcoma, a well-differentiated liposarcoma/atypical lipomatous tumor component adjacent to a high-grade sarcomatous component. (d) And a positive MDM2/CEP12 FISH assay in a dedifferentiated liposarcoma similar to the fluorescent photomicrograph in panel b.
(a) Spindle cell/pleomorphic lipoma, adipocytes with atypical nuclei admixed with ‘ropey’ collagen. (b) Polysomic MDM2/CEP12 FISH assay in a spindle cell/pleomorphic lipoma (same number of MDM2-red signals as CEP12-green signals). (c) Lipoma, adipocytes lacking atypia. (d) Lipoma with a negative MDM2/CEP12 FISH assay (two MDM2-red signals and two CEP12-green signals).
In pleomorphic sarcoma, not otherwise specified, MDM2 amplification was identified in 40% of (4/10) cases (MDM2/CEP12 ratio: 3.3, 2.3, 6.3, and 2.1), while 100% (10/10) were polysomic for CEP12 (average MDM2/CEP12 ratio: 1.6). Of the remainder of the neoplasms studied, extra-skeletal myxoid chondrosarcoma (n=10), myxofibrosarcoma (n=1), clear cell sarcomas (n=4), low-grade fibromyxoid sarcomas (n=10), solitary fibrous tumors/hemangiopericytomas (n=10), malignant melanoma (n=1), neuroblastoma (n=1), Ewing's sarcomas/primitive neuroectodermal tumors (n=3), desmoplastic small round cell tumor (n=1), alveolar rhabdomyosarcomas (n=3), gastrointestinal stromal tumors (n=4), leiomyosarcoma (n=1), and synovial sarcomas (n=4) displayed no evidence of MDM2 amplification or chromosome 12 polysomy. However, a small subset of these soft tissue neoplasms (5% or 3/63) did have evidence of MDM2 amplification, including one epithelioid sarcoma (n=1), one malignant peripheral nerve sheath tumor (n=4), and one myxoid liposarcoma (n=5, diagnosis confirmed by DDIT3 break-apart FISH).
Discussion
Well-differentiated liposarcoma/atypical lipomatous tumor is the most common type of liposarcoma, but can be difficult to distinguish from benign lipomatous tumors. Lipoblasts may not be present in well-differentiated liposarcoma/atypical lipomatous tumor, and alternatively, even benign lipomatous tumors such as spindle cell lipomas may have scattered lipoblasts. Enlarged hyperchromatic nuclei, the histologic feature most reliable for a diagnosis of well-differentiated liposarcoma/atypical lipomatous tumor, may be widely scattered and not even present in a small biopsy specimen. In addition, atypical nuclei are characteristically found in spindle cell/pleomorphic lipomas, a tumor that is difficult to distinguish from well-differentiated liposarcoma/atypical lipomatous tumor based solely on histological features. In our experience, one of the more common diagnostic dilemmas that arise in our consultation practice is the distinction of well-differentiated liposarcomas/atypical lipomatous tumors from benign lipomatous tumors. However, with the identification of characteristic ring and giant chromosomes largely comprised of 12q13-15 amplicons in well-differentiated liposarcomas/atypical lipomatous tumors, ancillary diagnostic tools can now be utilized to distinguish benign lipomatous tumors from well-differentiated liposarcomas/atypical lipomatous tumors.
Immunohistochemistry (IHC) is often the first ancillary tool performed in current day diagnostic pathology to differentiate tumors resembling one another morphologically. The 12q13-15 amplicons characteristic of well-differentiated liposarcomas/atypical lipomatous tumors and dedifferentiated liposarcomas have been shown to include both MDM2 and CDK4 genes. Therefore, one might expect their corresponding proteins to be overexpressed in these lesions as a result of their amplification, thereby facilitating this evaluation. To evaluate this possibility, Binh et al14 analyzed a large number of soft tissue tumors by IHC using antibodies to MDM2 and CDK4. In their hands, the sensitivity and specificity of MDM2 and CDK4 expression in distinguishing well-differentiated liposarcomas/atypical lipomatous tumors, and dedifferentiated liposarcomas from other soft tissue neoplasms was 97 and 92, and 83 and 95%, respectively. In their series, all 44 cases of well-differentiated liposarcomas/atypical lipomatous tumors were MDM2 positive, but 4.2% of benign lipomatous tumors and 18.9% of various sarcomas including malignant peripheral nerve sheath tumor, myxofibrosarcoma, embryonal rhabdomyosarcoma, malignant fibrous histiocytoma, leiomyosarcoma, myxoid liposarcoma, synovial sarcoma, angiosarcoma, dermatofibrosarcoma protuberans, clear cell sarcoma, Kaposi's sarcoma, epithelioid sarcoma, and desmoplastic small round cell tumor were also immunoreactive. Although there was decreased sensitivity when CDK4 immunostaining was added in combination, the specificity of this staining panel improved, as only 2.3% of benign lipomatous tumors and 3.7% of sarcomas were positive for both MDM2 and CDK4. Similar results have been reported from other laboratories with excellent interobserver concordance.15 However, as there is typically not an absolute correlation between gene amplification and protein overexpression, detection of the underlying genetic alteration itself might be expected to yield higher sensitivity and specificity when compared to IHC.
Although MDM2 FISH probes have been used in combination with other molecular techniques in numerous studies to define and demonstrate the 12q13-15 amplicons characteristic of well-differentiated liposarcoma/atypical lipomatous tumor and dedifferentiated liposarcoma, this tool has not been formally evaluated in the published literature as an ancillary test to help differentiate these entities from their morphological mimics.10, 16, 17, 18, 19 Jacob et al20 reported using a custom-designed FISH probe for MDM2 along with a commercially available CEP12 probe. Similar to our data, in their smaller series of lipomatous tumors, all benign lipomatous tumors lacked MDM2 amplification, while all well-differentiated liposarcoma/atypical lipomatous tumor exhibited amplification.
In this study, we have shown the MDM2/CEP12 FISH assay to be a sensitive and specific tool for well-differentiated liposarcoma/atypical lipomatous tumor and dedifferentiated liposarcoma, when compared to other lipomatous neoplasms. Thus, this technique provides an even greater sensitivity and specificity than has been previously achieved using immunohistochemical stains. All of the well-differentiated liposarcomas/atypical lipomatous tumors analyzed in this study were lipoma-like or sclerosing subtypes. However, the specificity does decrease when evaluating high-grade sarcomas, as MDM2 amplification was seen in a subset of high-grade sarcomas other than dedifferentiated liposarcomas. Interestingly, one of our myxoid liposarcomas (n=5) showed MDM2 amplification, while it also showed the evidence of the characteristic t(12;16), as demonstrated by break-apart FISH assays specific for DDIT3 (12q13 also known as CHOP) and FUS (16p11). The MDM2/CEP12 ratio was low in this neoplasm (3.1). Although we do not have a plausible explanation for the low-level MDM2 amplification in this neoplasm, loss of p53 in myxoid liposarcoma has been correlated with a worse prognosis, suggesting that MDM2 amplification could provide an alternate mechanism of affecting the p53 function in such cases.21 The single case of malignant peripheral nerve sheath tumor with MDM2 amplification (n=4) was from the retroperitoneum and showed divergent rhabdomyosarcomatous differentiation. The S-100 protein stain was only focally positive and definitive areas of well-differentiated liposarcoma/atypical lipomatous tumor, to suggest this could be part of a dedifferentiated liposarcoma, were not identified. Therefore, it is possible that this neoplasm could represent a dedifferentiated liposarcoma in which characteristic areas of well-differentiated liposarcoma/atypical lipomatous tumor were either not sampled or were overrun by the dedifferentiated component. The single example of epithelioid sarcoma in this study showed high-level MDM2 amplification. Additional evaluation of a larger number of epithelioid sarcomas will be needed to establish the true incidence of MDM2 amplification in this tumor type.
Pleomorphic sarcoma, not otherwise specified (malignant fibrous histiocytoma, nonmyxoid) tend to have complex karyotypes including extensive polysomy.22 MDM2 as well as other proto-oncogenes from the 12q13-15 chromosome region have been reported to be amplified in cases of pleomorphic sarcoma, not otherwise specified.22 Therefore, it was not a surprise to discover 40% (4/10) of the pleomorphic sarcomas from our study to be amplified for MDM2 and all (10/10) to be polysomic for CEP12. As the main differential diagnosis for dedifferentiated liposarcoma is pleomorphic sarcoma, not otherwise specified, and both entities display MDM2 amplification, the described MDM2 FISH assay is not as useful in this setting for distinguishing between these neoplasms. From a clinical standpoint, differentiation is not essential on needle core biopsy of a deep-seated mass, because both require the same surgical approach. Dedifferentiated liposarcoma and pleomorphic sarcoma, not otherwise specified, can only be differentiated on complete examination of the entire neoplasm by identifying a well-differentiated liposarcoma component in dedifferentiated liposarcomas. Further more, the high-grade sarcoma component of a dedifferentiated liposarcoma may overgrow the well-differentiated liposarcoma component becoming indistinguishable from a pleomorphic sarcoma, not otherwise specified. This phenomenon along with not sampling the well-differentiated liposarcoma component are plausible explanations for the MDM2 amplification detected in a percentage of our classified pleomorphic sarcoma, not otherwise specified cases. In fact, recent studies suggest that most pleomorphic sarcoma, not otherwise specified cases, otherwise known as malignant fibrous histiocytomas and their variants that arise in the retroperitoneum are actually dedifferentiated liposarcomas based on histological review, immunoprofile, and genomic profile.23, 24
In conclusion, the identification of MDM2 amplification by FISH is a useful ancillary tool in the diagnosis of well-differentiated liposarcomas/atypical lipomatous tumors, as none of the benign lesions commonly considered in its histologic differential diagnosis were found to harbor MDM2 amplification. Moreover, the nonatypical cells in well-differentiated liposarcomas/atypical lipomatous tumors showed MDM2 amplification, making the probe a valuable diagnostic tool even in small specimens where cytologic atypia is not identified but where well-differentiated liposarcoma/atypical lipomatous tumor remains a strong diagnostic consideration. While this study included only well-characterized tumor samples to illustrate the probe's performance, we predict that the MDM2 FISH assay will become a valuable tool in the evaluation of difficult lipomatous lesions.
References
Nascimento AG . Dedifferentiated liposarcoma. Semin Diagn Pathol 2001;18:263–266.
Laurino L, Furlanetto A, Orvieto E, et al. Well-differentiated liposarcoma (atypical lipomatous tumors). Semin Diagn Pathol 2001;18:258–262.
Weiss SW, Goldblum JR . Liposarcoma. Enzinger and Weiss's Soft Tissue Tumors, Vol., 5th edn. Mosby: St Louis, 2008, pp 477–516.
Enzinger FM, Winslow DJ . Liposarcoma. A study of 103 cases. Virchows Arch Pathol Anat Physiol Klin Med 1962;335:367–388.
Evans HL . Liposarcoma: a study of 55 cases with a reassessment of its classification. Am J Surg Pathol 1979;3:507–523.
Karakousis CP, Dal Cin P, Turc-Carel C, et al. Chromosomal changes in soft tissue sarcomas. A new diagnostic parameter. Arch Surg 1987;122:1257–1260.
Pedeutour F, Suijkerbuijk RF, Van Gaal J, et al. Chromosome 12 origin in rings and giant markers in well-differentiated liposarcoma. Cancer Genet Cytogenet 1993;66:133–134.
Dal Cin P, Kools P, Sciot R, et al. Cytogenetic and fluorescence in situ hybridization investigation of ring chromosomes characterizing a specific pathologic subgroup of adipose tissue tumors. Cancer Genet Cytogenet 1993;68:85–90.
Pedeutour F, Suijkerbuijk RF, Forus A, et al. Complex composition and co-amplification of SAS and MDM2 in ring and giant rod marker chromosomes in well-differentiated liposarcoma. Genes Chromosomes Cancer 1994;10:85–94.
Pedeutour F, Forus A, Coindre JM, et al. Structure of the supernumerary ring and giant rod chromosomes in adipose tissue tumors. Genes Chromosomes Cancer 1999;24:30–41.
Vargas DA, Takahashi S, Ronai Z . Mdm2: A regulator of cell growth and death. Adv Cancer Res 2003;89:1–34.
Skacel M, Skilton B, Pettay JD, et al. Tissue microarrays: a powerful tool for high-throughput analysis of clinical specimens: a review of the method with validation data. Appl Immunohistochem Mol Morphol 2002;10:1–6.
Patel RM, Downs-Kelly E, Weiss SW, et al. Dual-color, break-apart fluorescence in situ hybridization for EWS gene rearrangement distinguishes clear cell sarcoma of soft tissue from malignant melanoma. Mod Pathol 2005;18:1585–1590.
Binh MB, Sastre-Garau X, Guillou L, et al. MDM2 and CDK4 immunostainings are useful adjuncts in diagnosing well-differentiated and dedifferentiated liposarcoma subtypes: a comparative analysis of 559 soft tissue neoplasms with genetic data. Am J Surg Pathol 2005;29:1340–1347.
Binh MB, Garau XS, Guillou L, et al. Reproducibility of MDM2 and CDK4 staining in soft tissue tumors. Am J Clin Pathol 2006;125:693–697.
Shimada S, Ishizawa T, Ishizawa K, et al. The value of MDM2 and CDK4 amplification levels using real-time polymerase chain reaction for the differential diagnosis of liposarcomas and their histologic mimickers. Hum Pathol 2006;37:1123–1129.
Forus A, Bjerkehagen B, Sirvent N, et al. A well-differentiated liposarcoma with a new type of chromosome 12-derived markers. Cancer Genet Cytogenet 2001;131:13–18.
Micci F, Teixeira MR, Bjerkehagen B, et al. Characterization of supernumerary rings and giant marker chromosomes in well-differentiated lipomatous tumors by a combination of G-banding, CGH, M-FISH, and chromosome- and locus-specific FISH. Cytogenet Genome Res 2002;97:13–19.
Pilotti S, Della Torre G, Mezzelani A, et al. The expression of MDM2/CDK4 gene product in the differential diagnosis of well-differentiated liposarcoma and large deep-seated lipoma. Br J Cancer 2000;82:1271–1275.
Jacob EK, Erickson-Johnson M, Wang X, et al. Assessment of MDM2 amplification using fluorescence in situ hybridization on paraffin embedded tissues discriminates atypical lipomatous tumors from lipomas. Mod Pathol 2006;19:13A.
Antonescu CR, Tschernyavsky SJ, Decuseara R, et al. Prognostic impact of P53 status, TLS-CHOP fusion transcript structure, and histological grade in myxoid liposarcoma: a molecular and clinicopathologic study of 82 cases. Clin Cancer Res 2001;7:3977–3987.
Fletcher CDM, van den Berg E, Molenaar WM . Pleomorphic malignant fibrous histiocytoma/Undifferentiated high grade pleomorphic sarcoma In: Fletcher CDM, Krishnan Unni K, Mertens F (eds). World Health Organization Classification of Tumours Pathology and Genetics Tumours of Soft Tissue and Bone, Vol. IARC Press: Lyon, 2002, pp 120–122.
Coindre JM, Hostein I, Maire G, et al. Inflammatory malignant fibrous histiocytomas and dedifferentiated liposarcomas: histological review, genomic profile, and MDM2 and CDK4 status favour a single entity. J Pathol 2004;203:822–830.
Coindre JM, Mariani O, Chibon F, et al. Most malignant fibrous histiocytomas developed in the retroperitoneum are dedifferentiated liposarcomas: a review of 25 cases initially diagnosed as malignant fibrous histiocytoma. Mod Pathol 2003;16:256–262.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Weaver, J., Downs-Kelly, E., Goldblum, J. et al. Fluorescence in situ hybridization for MDM2 gene amplification as a diagnostic tool in lipomatous neoplasms. Mod Pathol 21, 943–949 (2008). https://doi.org/10.1038/modpathol.2008.84
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2008.84
Keywords
This article is cited by
-
Primary cardiac sarcoma: demographics, genomic study correlation, and survival benefits of surgery with adjuvant therapy in U.S. population
Clinical Research in Cardiology (2024)
-
Nodular cystic fat necrosis: a distinctive rare soft-tissue mass
Skeletal Radiology (2024)
-
Diagnostic value of MDM2 RNA in situ hybridization for low-grade osteosarcoma: Consistency comparison of RNA in situ hybridization, fluorescence in situ hybridization, and immunohistochemistry
Virchows Archiv (2023)
-
Accuracy of core needle biopsy for histologic diagnosis of soft tissue sarcoma
Scientific Reports (2022)
-
Perinephric myxoid pseudotumor of fat: a multimodality imaging case series
Abdominal Radiology (2022)