Abstract
Despite recent advances in surgical and multidisciplinary treatment, prognosis for patients with esophageal adenocarcinoma remains poor, and the low prognostic significance of pTNM staging suggests that additional parameters are needed. To identify genomic abnormalities characteristic of esophageal adenocarcinoma, a panel of 33 samples obtained at surgery from previously untreated patients were analyzed by muliplex ligation-dependent probe amplification technique. We detected frequent gains of 6p, 8q, 13q, 17q, 20q, and losses of 4q, 5q, 15q, and 18q. When DNA copy number changes were correlated to clinicopathological features of patients no association was found between the number of chromosomal aberrations and gender, age, tumor grade or pTNM staging. However, interestingly, a significant correlation between patient survival and total number of chromosomal aberrations was found when esophageal adenocarcinoma cases were stratified according to the median of survival (20 months) (P=0.002) or the median of aberrations (12 aberrations) (P=0.014). Evaluation of the distribution of gains and losses at the level of single chromosomes indicated that gains on chromosomes 5, 6, 8, 11, 20 and losses on chromosomes 1, 3, 5, 11, and 18 were significantly different in the two survival groups. Furthermore, when single gene imbalances were analyzed in further details, we found that besides alterations that involve genes shared by both survival groups, a few genes (KIAA0170, EMS1, ABCC4, F3, and MIF) were altered only in samples from patients with poor survival. Thus, we established a good correlation between the total number of chromosomal alterations and survival, suggesting that the estimation of total imbalances might represent an additional indicator of disease outcome. In addition, the finding of alterations specific for the more aggressive esophageal adenocarcinoma subset might represent promising biomarkers to increase the accuracy of clinical outcome prediction.
Similar content being viewed by others
Main
On the basis of histological characteristics, esophageal carcinoma can be subdivided into two major types: esophageal squamous cell carcinoma and esophageal adenocarcinoma. Over the past few decades, although the incidence of the former has remained stable, the incidence of esophageal adenocarcinoma has risen in the western world and Asia.1, 2, 3
Despite recent advances in therapies, including surgery, chemotherapy, radiotherapy, immunotherapy, and combinations thereof, the prognosis for patients with esophageal adenocarcinoma remains poor. Tumor recurrence frequently occurs within 2 years, although some patients survive long after surgery; a considerable variability and heterogeneity within the tumor stages exists, but none of the staging criteria accurately predicts the prognosis for an individual patient.
Many studies have attempted to clarify the molecular mechanisms underlying the biological behavior of esophageal adenocarcinoma. A number of genetic alterations have been observed, gain of chromosome 20q being the most recurrent. Gains were also frequently observed on chromosomes 6q, 7p, 7q, 8q, 11q, 15q, and 17q; recurrent losses were described on chromosomes 1p, 3p, 4p, 4q, 5q, 8p, 9p, 17p, and 18q.4, 5, 6, 7, 8 The majority of these studies used conventional comparative genomic hybridization (chromosomal-CGH), making identification of the genes involved difficult; to date, the genetic alterations associated with disease onset and progression remain largely unknown.
The multiplex ligation-dependent probe amplification (MLPA) technique, which allows detection of single gene alterations, fills the gap between techniques that provide information on large chromosomal region but are time-consuming and require relative large DNA amounts (chromosomal-CGH) and techniques that are faster and need smaller DNA amounts but provide information only on specific loci (LOH, quantitative PCR). In terms of robustness, MLPA has been compared to array-CGH,9 and represents an efficient method for simultaneous screening of copy number imbalance in multiple genomic regions while maintaining a single gene resolution. Thus, MLPA technique provides a promising starting point for the identification of novel candidate genes affected by genomic abnormalities.
In this paper, we analyzed 33 esophageal adenocarcinoma samples obtained at surgery from previously untreated patients, and found a significant correlation between patient survival and total number of chromosomal aberrations. Interestingly, some of the detected genetic imbalances appeared to be specific for the poorest survival group, thus highlighting the possibility of defining new prognostic biomarkers.
Materials and methods
Patients and Controls
A total of 33 patients diagnosed with esophageal adenocarcinoma between 1990 and 2000 gave their informed consent to the study (27 men and 6 women, mean age 66.5 years, range 28–85 years); their clinical and histopathologic characteristics are summarized in Table 1. All patients underwent esophagectomy without preoperative radio- or chemotherapy, and were staged according to the International Union Against Cancer TNM classification; follow-up data were available for all cases (mean follow-up 37 months, range 1–114). All analyses were performed on DNA extracted from the original tumor sample obtained at surgery and stored at −80°C until use. The presence of high neoplastic content (>70% tumor tissue) was assessed by histological analysis. DNA obtained from eight controls (five men and three women) was also used; six samples were frozen tissues of nonneoplastic mucosa from the margins of resection specimens and two were samples of peripheral blood mononuclear cells (PBMC) from healthy volunteers.
DNA Extraction and MLPA Analysis
DNA was extracted using the phenol–chloroform method.10 The concentration was quantified spectrophotometrically and the DNA quality was checked on a 1% agarose gel. By means of MLPA up to 45 different sequences can be targeted in a single, semiquantitative PCR-based experiment.11 Human chromosomal aberration test kits (SALSA kits P005, P006, and P007) were used according to manufacturer's instructions (MRC Holland, The Netherlands). Altogether, these kits cover 112 genes, some of which were rejected due to their variability in normal control samples (see below). Amplification products were separated by capillary electrophoresis on an ABI 310 sequencer (Applied Biosystems, CA, USA). As amplification control, three samples exhibiting amplification of the BRCA2 gene and one with gain of the BRCA1 gene were also analyzed with the MLPA P045 and P087 kits (MRC Holland), specific for the entire BRCA2 and BRCA1 genes, respectively.12, 13
In the data normalization procedure, the relative peak area for each probe was obtained as fractions of the total sum of peak areas for a given sample. Then, the resulting relative peak areas were divided by the average of relative peak areas derived from normal control samples; in the absence of amplification or loss, the ratio is expected equal to 1, whereas higher or lower values indicate gains or losses. Highly amplified genes (normalized copy number >2) were left out when calculating the total sum of peak areas, and the peak areas were renormalized.
Data were also analyzed and normalized by two other distinct ways in order to work out experimental variability, noise, and known drawbacks of MLPA kits. At first, the MRC-Holland software Coffalyser version 2 was used to correct slope where necessary, and to detect gains and losses in gene copy number. Next, as a large number of chromosomal abnormalities may be present in cancer cells, we also normalized the data by selecting internal control probes whose pattern of amplification did not deviate from that of the reference controls, ie genes with the lowest relative peak area variation in all experiments.
A difference between control and tumor samples was interpreted as significant only when the ratio was less than 0.7 (loss) or higher than 1.3 (gain). Only gains and losses that were concordant using these different approaches were considered reliable.
Fluorescence In Situ Hybridization Analysis
Fluorescence in situ hybridization (FISH) was performed on 4-μm paraffin-embedded sections. After two cycles of deparaffinization in xylene for 15 min and dehydration in 100% ethanol for 1 min, the samples were permeabilized by microwave oven with citrate buffer 0.01 M (pH 6.0) for 5 min, and then digested with proteinase K (25 μg/ml) in 50 mM Tris HCl pH 7.6/1 mM CaCl2, at 37°C for 15 min. Sections were then hybridized overnight at 37°C with LSI TOP2A SpectrumOrange, ERBB2 Spectrumgreen, and CEP17 SpectrumAqua probes (Vysis-Abbott, IL, USA), according to manufacturer's instructions; before hybridization, probes and target DNA were codenatured using Hybrite system (Vysis) at 80°C for 10 min. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and mounted in antifade solution Vectashield (Vector, UK). Analysis was carried out with a fluorescence microscope (Zeiss Axioskop 2 Plus, Zeiss, Germany) equipped with single-band filters. For each probe set, 60 nonoverlapping nuclei were enumerated and results were reported as the ratio between number of signals ERBB2/CEP17 and TOP2/CEP17. The cutoff value for designation of amplification was a ratio ≥2 for each probe.
Statistical Analysis
Association between DNA copy number alterations and survival was analyzed using the Mann–Whitney U-test; the same test was performed on specific subset of chromosomes and genes. Differences with a P-value<0.05 were considered statistically significant. Statistical analysis was performed by using the StatGraphics software, version 2.6. Survival curves and probabilities were estimated using the Kaplan–Meier analysis.
Results
Amplification Pattern in Control Samples
To determine the reliability of the MLPA kits employed and to establish the relative pattern of probe amplification in normal control samples, we analyzed eight nonneoplastic DNA, six obtained from normal esophageal mucosa distant from the margin of the resected tumor, and two from PBMC. In all controls the relative areas of amplification for most probes were homogeneous, with standard deviations ranging between ±0.03 and ±0.1 and considered compatible with experimental variability declared by the manufacturer. Probes for IL12A and EHF genes (SALSA P006 kit) and CTPS probe (SALSA P007 kit) that exhibited a variability higher than 10% were not considered in the study as well as probes for X and Y chromosomes. Therefore, the final analysis was performed on 103 rather than 112 genes.
Association Between Copy Number Alterations and Survival in Esophageal Adenocarcinoma Cases
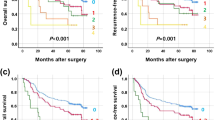
Results of the MLPA analysis revealed DNA copy number changes in all samples (mean 15.6; range 3–33). Altogether 97 (94%) out of 103 genes examined presented alterations and at least one gene imbalance was present in almost all chromosomes. When the number of chromosomal aberrations were correlated to clinicopathological features no association was found between the number of alterations and patients gender, age, tumor grade, or pTNM staging (Table 1). However, when the total number of chromosomal imbalances was correlated to patient survival, a significant association (P=0.002) between the total number of gains/losses and survival was found. In particular, by dividing the patients according to their median survival time (20 months), we found that the mean number of quantitatively altered genes per tumor was 20.7 (range 6–33) in patients with a survival ≤20 months (17 patients), compared to 10.1 (range 3–22) in patients with a survival >20 months (16 patients) (Table 2). Similarly, stratification of the patients according to the median number of chromosomal imbalances (12 aberrations), showed a good association with survival (P=0.014). Indeed, the 17 patients with ≤12 alterations had a mean survival of 51 months (range 3–114), whereas the 16 patients with >12 aberrations showed a lower survival (mean 23 months, range 4–78). The correlation between the number of chromosomal abnormalities and survival was also significant when analysis was restricted to the 19 patients classified in the same pT3N1M0 stage. Among them, the 9 patients exhibiting a median number of aberration ≤12 showed a mean survival of 37 months (range 9–96), whereas the mean survival of the 10 patients exhibiting >12 aberrations was 11 months (range 4–20) (P=0.003). Reverse association between survival and DNA copy number alterations was also sustained by Kaplan–Meier survival analysis, as reported in Figure 1.
Analysis of Chromosomal Regions Involved in DNA Copy Number Alterations
Thus we analyzed single gene imbalance in further details. Applying a cutoff level of 20% frequency, we found an imbalance of 32 genes (26 gains, 8 losses) in the ≤20-month survival group and of 7 genes (6 gains, 1 loss) in the >20-month survival group. Both subgroups showed amplifications on chromosomes 6p12, 8q24, 13q34, 17q11–q21, 20q12–q13, whereas losses where more frequent on chromosomes 4q22, 15q21–q22, and 18q21.3. However, when the frequency of total gains and losses was compared at single chromosome level in the two subgroups, we found a statistically different number of gains on chromosomes 5, 6, 7, 11, and 20 (P<0.05), whereas chromosome 8 showed a P=0.055; losses were significant for chromosomes 1, 3, 5, 11 (P<0.03), and chromosome 18 showed a P=0.056 (Table 3). Nevertheless, when the analysis was restricted to specific chromosomal regions, we found that the difference between the two groups was even higher for chromosomes 11q13 and 20q13, and that imbalances on chromosomes 8q24.3 and 18q21.3 became statistically significant (Table 3).
As expected, comparison of the specific alterations present in the two survival groups revealed an overlap of some genetic changes (Table 4). Common genes exhibiting the same high frequency of DNA copy number gains were VEGF (6p12; >50%), PTK2 (8q24; >55%), ING1 (13q34; >25%), and SCYA3 (17q11–q21; >69%). Genes that were altered in both groups but with different frequencies were NCOA3 (20q12; 59% in the poorest survival group vs 37% in the >20 months survival group) and PTPN1 (20q13.13; 53 vs 25%). Genes that were amplified in both groups but only in a few patients includes BAK1 (6p21.3), ABCB1 (7q21.1), PRKDC (8q11), all with frequencies around 20%. Interestingly, this analysis showed that KIAA0170 (6p21.31), EMS1 (11q13), and ABCC4 (13q32) genes were amplified with frequencies between 23 and 35% only in the group with poor survival.
A similar behavior was observed for losses (Table 4), with genes ABCG2 (4q22; >20%), DCC (18q21.3; >30%), exhibiting comparable frequencies of loss in both subgroups; genes F3 (1p22–p21; 35%) and MIF (22q11.23; 23%) were instead lost only in the group with poor survival. No specific alterations were found in the >20-month survival group. Thus, although the difference between single gene did not reach statistical significance, some of the observed alterations seem to be a unique feature of rapid progressing patients.
Interestingly, in samples from patients with poor survival we found regions of imbalance at chromosomes 8q24.3, 11q13, 20q13.1, and 18q21.3, characterized by the concurrent amplification or loss of at least two contiguous probes (Table 5).
MLPA Data Validation
Data obtained with MLPA kits were validated by both FISH analysis and gene-specific MLPA. FISH validation of DNA copy number gains was carried out on two esophageal adenocarcinoma samples which showed ERBB2 and TOPO-2 amplification by MLPA analysis. Locus-specific probes for ERBB2 and TOPO-2 were co-hybridized to paraffin-embedded tissue sections corresponding to the frozen biopsies used for DNA extraction and MLPA experiments. Both samples resulted amplified for ERBB2 and TOPO-2 genes. As shown in Figure 2, one case showed high-grade amplification of both genes (>20 copies) with a cluster of co-localized signals inside the nuclei (Figure 2). The second case showed a moderate amplification (8–10 signals for TOPO-2 and 5–7 for ERBB2), with a diffuse pattern of signals (data not shown). Interestingly, the different levels of amplification found by FISH analysis confirmed the data obtained by MLPA, where the ratio for both genes was ≥2 in the first case, and 1.5 (TOPO-2) and 1.3 (ERBB2) in the second one.
In an independent series of experiments, we also found a good concordance in analyzing three samples that showed gains of BRCA2 gene and one exhibiting amplification of BRCA1 gene, using MLPA kits specific for the whole genes (P045 and P087, respectively). Amplifications of BRCA1 and BRCA2, as well as the sequences just upstream and downstream of the latter gene, were confirmed to be amplified in all cases (data not shown). Altogether, these results are in excellent concordance with those obtained with P005, P006, and P007 MLPA kits.
Discussion
Identifying genetic and/or biological features of tumors that correlate with clinical characteristics is an important goal in cancer research. Nonetheless, although molecular research has generated an enormous amount of information on the progression processes involved in many tumor types, to date very little is known regarding esophageal adenocarcinoma. Studies carried out by classical CGH have demonstrated multiple chromosomal copy number changes, including gains in chromosomes 6q, 7p, 7q, 8q, 11q, 15q, 17q, and 20q, as well as losses at chromosomes 1p, 3p, 4p, 4q, 5q, 8p, 9p, 17p, and 18q.4, 5, 6, 7, 8, 14 However, the minimal amplified region detected by conventional CGH usually spans several megabases, making it difficult to identify specific candidate genes involved in tumor development.
The present study reports an analysis carried out to identify DNA copy number changes and putative genes of interest that correlate with clinicopathologic features of 33 esophageal adenocarcinoma samples using the MLPA technique. Applying this technique, which is comparable to array-CGH in terms of robustness,9 we not only found chromosomal gains and losses that are largely in agreement with the literature, but were also able to identify specific genes that are located within the candidate chromosomal region. Although no correlation between gender, age, grade, or pTNM stage could be found, we observed a highly significant association between the number of gains/losses and survival. Indeed, by dividing the 33 esophageal adenocarcinoma samples into two subgroups based on the median survival (≤20 months, 17 cases and >20 months, 16 cases), we found a statistically significant difference in the total number of imbalances (P=0.002). Consistently, stratification of the samples by the median number of chromosomal alterations (≤12 aberrations, 17 cases and >12 aberrations, 16 cases), revealed that the two groups differed significantly in their overall survival (P=0.014). Albeit the increase in copy number abnormalities observed in patients with a worse outcome could simply reflect a general increase in genetic instability often detected in tumor progression, we also observed a highly significant association between the number of abnormalities and survival in patients with tumors diagnosed at the same pT3N1M0 staging (Figure 1).
Analysis of the genes involved revealed that, besides genes affected in both subgroups, some genes, such as KIAA0170 (6p21.31), EMS1 (11q13), ABCC4 (13q32), F3 (1p22–p21) and MIF (22q11.23) were affected only in the poor survival group. Furthermore, we found regions of imbalance, characterized by the concomitant gain or loss of two contiguous genes, present exclusively or with much higher frequency in samples from patients with a survival ≤20 months.
Recurrent DNA gains on chromosome 6p have been reported in esophageal adenocarcinoma,6, 7, 15 but to date they have not been well characterized. Our analysis revealed that besides VEGF amplification (6p12) common to 50% of esophageal adenocarcinoma samples, other alterations were found exclusively or mostly in the ≤20-month survival subgroup, such as gain of KIAA0170 and IER3 genes at 6p21.3. In addition, at the 8q24.3 region we were able to identify one putative area of amplification of 3.2 Mbp that appears specific for samples from patients with worst survival. In the poor survival subgroup, we also observed at chromosome 11q13 two areas of gain marked by CCND1, FGF3, and EMS1 genes. Interestingly, all these genes have been associated with different tumors;16, 17, 18 in particular, EMS1 amplification and overexpression was related to poor prognosis in breast cancer19 and disease progression in esophageal squamous cell carcinoma.20
A DNA gain at chromosome 20q is the most frequent chromosomal aberration described in esophageal adenocarcinoma, being found in up to 60% of cases.15 Albrecht et al,21 using array-CGH reported ZNF217 (20q13.2) and MYBL2 (20q13.12) amplifications in 39% of the cases. We also found amplifications at chromosome 20q13; however, in our study the sequences used were specific for NCOA3 and PTPN1 genes that are located in a region internal to that delimited by ZNF217 and MYBL2. Both NCOA3 and PTPN1 genes exhibited an high-gain frequency in all esophageal adenocarcinoma samples; however, they were coamplified in 44% of patients with lower survival and in only 6% of those with survival >20 months. In agreement with Albrecht's data,21 we did not find significant alterations of STK15 gene (20q13.2), often amplified in ovarian carcinoma.
Although DNA copy number changes cannot directly be used to predict RNA transcript levels, several recent studies have shown a significant correlation between somatic DNA alterations and the corresponding profile of RNA changes.20, 22, 23 Interestingly, within chromosome 15q26, we found recurrent DNA sequence copy number gains for IGF1R in both survival subgroups even though with different frequencies (35% in the ≤20-month survival group vs 12% in the good survival group). This result confirms previous studies by Iravani et al,24 who reported a correlation between IGF1R protein expression and progression of esophageal adenocarcinoma.
In conclusion, we found a good correlation between the total number of chromosomal alterations and survival of esophageal adenocarcinoma patients, suggesting that the estimation of the total number of imbalances might represent an additional indicator of disease outcome. Furthermore, although further studies are warranted to validate the observed alterations, some of them appear to be specific for the more aggressive esophageal adenocarcinoma subset, and therefore might represent useful biomarkers to increase the accuracy of clinical outcome prediction.
References
Bollschweiler E, Wolfgarten E, Gutschow C, et al. Demographic variations in the rising incidence of esophageal adenocarcinoma in white males. Cancer 2001;92:549–555.
Devesa SS, Blot WJ, Fraumeni Jr JF . Changing patterns in the incidence of esophageal and gastric carcinoma in the United States. Cancer 1998;83:2049–2053.
Vizcaino AP, Moreno V, Lambert R, et al. Time trends incidence of both major histologic types of esophageal carcinomas in selected countries, 1973–1995. Int J Cancer 2002;99:860–868.
Barrett MT, Galipeau PC, Sanchez CA, et al. Determination of the frequency of loss of heterozygosity in esophageal adenocarcinoma by cell sorting, whole genome amplification and microsatellite polymorphisms. Oncogene 1996;12:1873–1878.
Hage M, Siersema PD, Vissers KJ, et al. Genomic analysis of Barrett's esophagus after ablative therapy: persistence of genetic alterations at tumor suppressor loci. Int J Cancer 2006;118:155–160.
van Dekken H, Wink JC, Vissers KJ, et al. Genomic analysis of early adenocarcinoma of the esophagus or gastroesophageal junction: tumor progression is associated with alteration of 1q and 8p sequences. Genes Chromosomes Cancer 2006;45:516–525.
Riegman PH, Vissers KJ, Alers JC, et al. Genomic alterations in malignant transformation of Barrett's esophagus. Cancer Res 2001;61:3164–3170.
Koppert LB, Wijnhoven BP, van Dekken H, et al. The molecular biology of esophageal adenocarcinoma. J Surg Oncol 2005;92:169–190.
Kriek M, Knijnenburg J, White SJ, et al. Diagnosis of genetic abnormalities in developmentally delayed patients: a new strategy combining MLPA and array-CGH. Am J Med Genet 2007;143:610–614.
D'Andrea E, Saggioro D, Fleissner E, et al. Abelson murine leukemia virus-induced thymic lymphomas: transformation of a primitive lymphoid precursor. J Natl Cancer Inst 1987;79:189–195.
Schouten JP, McElgunn CJ, Waaijer R, et al. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res 2002;30:e57.
Montagna M, Dalla Palma M, Menin C, et al. Genomic rearrangements account for more than one-third of the BRCA1 mutations in northern Italian breast/ovarian cancer families. Hum Mol Genet 2003;12:1055–1061.
Agata S, Dalla Palma M, Callegaro M, et al. Large genomic deletions inactivate the BRCA2 gene in breast cancer families. J Med Genet 2005;42:e64.
Lin L, Wang Z, Prescott MS, et al. Multiple forms of genetic instability within a 2-Mb chromosomal segment of 3q26.3–q27 are associated with development of esophageal adenocarcinoma. Genes Chromosomes Cancer 2006;45:319–331.
Walch AK, Zitzelsberger HF, Bruch J, et al. Chromosomal imbalances in Barrett's adenocarcinoma and the metaplasia–dysplasia–carcinoma sequence. Am J Pathol 2000;156:555–566.
Fournier G, Latil A, Amet Y, et al. Gene amplifications in advanced-stage human prostate cancer. Urol Res 1995;22:343–347.
Hui R, Campbell DH, Lee CS, et al. EMS1 amplification can occur independently of CCND1 or INT-2 amplification at 11q13 and may identify different phenotypes in primary breast cancer. Oncogene 1997;15:1617–1623.
Hui R, Ball JR, Macmillan RD, et al. EMS1 gene expression in primary breast cancer: relationship to cyclin D1 and oestrogen receptor expression and patient survival. Oncogene 1998;17:1053–1059.
Rodrigo JP, Garcia LA, Ramos S, et al. EMS1 gene amplification correlates with poor prognosis in squamous cell carcinomas of the head and neck. Clin Cancer Res 2000;6:3177–3182.
Luo ML, Shen XM, Zhang Y, et al. Amplification and overexpression of CTTN (EMS1) contribute to the metastasis of esophageal squamous cell carcinoma by promoting cell migration and anoikis resistance. Cancer Res 2006;66:11690–11699.
Albrecht B, Hausmann M, Zitzelsberger H, et al. Array-based comparative genomic hybridization for the detection of DNA sequence copy number changes in Barrett's adenocarcinoma. J Pathol 2004;203:780–788.
van Dekken H, Vissers K, Tilanus HW, et al. Genomic array and expression analysis of frequent high-level amplifications in adenocarcinomas of the gastro-esophageal junction. Cancer Genet Cytogenet 2006;166:157–162.
Rodriguez V, Chen Y, Elkahloun A, et al. Chromosome 8 BAC array comparative genomic hybridization and expression analysis identify amplification and overexpression of TRMT12 in breast cancer. Genes Chromosomes Cancer 2007;46:694–707.
Iravani S, Zhang HQ, Yuan ZQ, et al. Modification of insulin-like growth factor 1 receptor, c-Src, and Bcl-XL protein expression during the progression of Barrett's neoplasia. Hum Pathol 2003;34:975–982.
Acknowledgements
This work was supported in part by grants from Italian Association for Research on Cancer (AIRC); Italian Foundation for Research on Cancer (FIRC); Ministry of University and Research; Ministry of Health; Regione Veneto (Ricerca Sanitaria Finalizzata); Fondazione Cassa di Risparmio di Padova e Rovigo. We thank Pierantonio Gallo and Renzo Grancara for artwork.
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclosure/conflict of interest
The authors have no potential conflict of interest to declare.
Rights and permissions
About this article
Cite this article
Pasello, G., Agata, S., Bonaldi, L. et al. DNA copy number alterations correlate with survival of esophageal adenocarcinoma patients. Mod Pathol 22, 58–65 (2009). https://doi.org/10.1038/modpathol.2008.150
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2008.150
Keywords
This article is cited by
-
Disseminated tumour cells with highly aberrant genomes are linked to poor prognosis in operable oesophageal adenocarcinoma
British Journal of Cancer (2017)
-
Integrated molecular analysis reveals complex interactions between genomic and epigenomic alterations in esophageal adenocarcinomas
Scientific Reports (2017)
-
Replication stress links structural and numerical cancer chromosomal instability
Nature (2013)
-
Growth inhibition, morphology change, and cell cycle alterations in NFBD1-depleted human esophageal cancer cells
Molecular and Cellular Biochemistry (2010)
-
Characterization of candidate gene copy number alterations in the 11q13 region along with BRAF and NRAS mutations in human melanoma
Modern Pathology (2009)