Abstract
Extraskeletal myxoid chondrosarcoma is a rare soft tissue tumor characterized by a nodular growth pattern with eosinophilic cells usually in a reticular pattern and abundant myxoid stroma. In contrast to other myxoid sarcomas, the majority of extraskeletal myxoid chondrosarcomas harbor a balanced translocation, t(9;22)(q22;q12), that fuses EWSR1 with NR4A3 (also known as CHN). Other less common translocations involving NR4A3 have also been described. We examined the diagnostic utility of fluorescence in situ hybridization for extraskeletal myxoid chondrosarcoma using the LSI EWSR1 break-apart probe (Abbott Molecular/Vysis, Des Plaines, IL, USA). Sixteen cases of extraskeletal myxoid chondrosarcoma with formalin-fixed paraffin-embedded tissue available were retrieved (1991–2007). The mean age at time of presentation was 57 years (range, 30–78). The male to female ratio was 7:1. All cases where either consistent with or highly suggestive of the diagnosis, with most of the primary tumors occurring in the thigh, inguinal or gluteal region. Fifteen of 16 cases were analyzable, of which 14 (93%) were positive for the rearrangement of the EWSR1 locus. In this study, the vast majority of extraskeletal myxoid chondrosarcomas are associated with a rearrangement at the EWSR1 locus (22q12). Fluorescence in situ hybridization is useful to support the diagnosis of extraskeletal myxoid chondrosarcomas and may help to differentiate it from mimics such as other myxoid sarcomas, particularly in limited biopsies.
Similar content being viewed by others
Main
Extraskeletal myxoid chondrosarcoma is a rare soft tissue tumor of uncertain origin that accounts for fewer than 3% of all soft tissue sarcomas.1 It most often occurs in middle-aged to elderly adults and is more common in males (ratio 2:1).1, 2 Extraskeletal myxoid chondrosarcoma typically arises in the deep soft tissue of the proximal extremities, most commonly in the thigh.1, 2 Although slow growing, these tumors have a significant risk of eventual relapse and metastases, especially pulmonary.1, 2, 3, 4 Altogether, 10-year survival has been recently estimated to be from 63 to 88%, with a reported 10-year disease-free survival between 14 and 36%.2, 4, 5, 6
Histologically, extraskeletal myxoid chondrosarcomas are multinodular and composed of polygonal to spindled eosinophilic cells in a typically prominent myxoid background. The tumor cells are characteristically arranged in a cord-like to reticular pattern but can have cribriform, solid and clustered arrangements. Cellular variants with higher-grade appearing features including diffuse sheets of mitotically active anaplastic epithelioid cells, and minimal myxoid matrix have also been described.1, 2, 3 Because of its myxoid stroma and varied morphological appearance, extraskeletal myxoid chondrosarcomas can be confused with other myxoid neoplasms, including chordomas, myxoid liposarcomas, myxomas, low-grade fibromyxoid sarcomas and myxofibrosarcomas, especially in small biopsy samples.2, 7
Immunohistochemical studies have generally not been helpful in establishing a diagnosis but they may help exclude other entities. Vimentin is the only marker consistently positive in extraskeletal myxoid chondrosarcomas and certainly does not convey specificity. Focal staining for cytokeratins and epithelial membrane antigen has been described.2 In contrast to other cartilaginous neoplasms, only a few extraskeletal myxoid chondrosarcomas actually express S-100 protein and often with focal to weak staining.2, 8 This finding, coupled with the fact that the vast majority of extraskeletal myxoid chondrosarcomas lack true cartilage and cartilage-specific matrix proteins, has raised questions whether extraskeletal myxoid chondrosarcomas are truly chondroid neoplasms as their name implies.1, 8, 9 Recent studies have also demonstrated that some extraskeletal myxoid chondrosarcomas may have neuroendocrine differentiation by immunohistochemical and ultrastructural studies.1, 2, 10, 11 As a result, these tumors are currently regarded as mesenchymal tumors of uncertain origin.1, 2
Unlike other myxoid sarcomas, extraskeletal myxoid chondrosarcomas are characterized with a balanced translocation involving the NR4A3 gene (also known as CHN, TEC, NOR1 or MINOR) located on 9q22. The most common fusion partner is the EWSR1 gene on 22q11.1, 2, 7, 11, 12 Other less frequently reported fusion partners include the TAF15 (also known as RBP56, hTAFii68 or TAF2N) on 17q11, TCF12 on 15q21 and, more recently, TFG on 3q12.1, 2, 7, 11, 13, 14 Although these translocations can be detected using reverse transcription–polymerase chain reaction,12, 15, 16, 17, 18 this molecular diagnostic assay is not readily accessible to most pathology practices.
Given the reported prevalence of EWSR1 involvement in extraskeletal myxoid chondrosarcomas, we examined the utility of fluorescence in situ hybridization using a commercially available EWSR1 break-apart probe as an ancillary diagnostic tool for extraskeletal myxoid chondrosarcoma.
Materials and methods
With institutional review board approval, 16 cases of extraskeletal myxoid chondrosarcoma with formalin-fixed paraffin-embedded tissue available were retrieved from the pathology files of The University of Texas MD Anderson Cancer Center from 1991 to 2007. Hematoxylin and eosin slides of all cases were reviewed by two soft tissue pathologists at this institution (WLW and AJL). Sections (4-μm thick) from representative blocks of tumor (metastatic or primary tumors) were prepared on positively charged slides. Tumors from a variety of specimen types were tested, including resections (seven), excisional biopsies (five), core biopsies (three) and cell block (one).
Fluorescence in situ hybridization for EWSR1 rearrangement was performed using the LSI EWSR1 dual-color, break-apart probe (Abbott Molecular/Vysis, Des Plaines, IL, USA) according to the manufacturer's recommendations. Tissue sections (4-μm thick) were placed onto slides, air-dried, and baked overnight at 60°C. Slides were deparaffinized in CitriSolv (Fisher, Vernon Hills, IL, USA) three times for 5 min and then immersed in 100% ethanol twice for 1 min. After air-drying, slides were treated in Paraffin Pretreatment solution (Paraffin Pretreatment Kit II; Abbott Molecular/Vysis) for 10 min at 80°C, washed with purified water for 3 min at room temperature, and treated with protease solution for 15 min, at 37°C. Slides were subsequently rinsed in purified water for 3 min, air-dried, and put in 2 × saline-sodium citrate buffer at 37°C for 30 min, dehydrated in 70, 85, and 100% ethanol, respectively, and allowed to air-dry. Next, 10 to 20 μl of LSI EWSR1 dual-color break-apart probe (Abbott Molecular/Vysis) was applied to the slides in an approximately 1 cm2 area selected for a pure tumor population (>90% tumor cells), and hybridization was performed at 37°C overnight in a moist chamber. Excess probe was washed away using 2 × saline-sodium citrate buffer/0.3% NP-40 (Fisher) at 73°C for 2 min, and the nuclei were counterstained with 4′,6-diamidino-2-phenylindole dihydrochloride/Vectashield (Vector Laboratories, Burlingame, CA, USA).
For scoring, the tissue sections were examined under a Zeiss fluorescence microscope (Carl Zeiss, Thornwood, NY, USA) using a × 10 ocular and × 63 and × 100 oil immersion lenses. The slides were scanned and images captured by a cytogenetic technologist and interpreted by a cytogeneticist, who estimated the percent positive nuclei. Only cell nuclei with one yellow (fusion), one green, and one red signal detected simultaneously were considered positive for EWSR1 rearrangement. Signals were considered to be colocalized when their distance was equal to or smaller than the size of the hybridization signal. Negative control values were established on cultures of bone marrow aspirate specimens from 10 known negative patients. The probe specificity was confirmed by mapping back to banded metaphase nuclei, and by hybridization to previously identified cases with rearrangements involving 22q12 (EWSR1) on conventional cytogenetic and fluorescence in situ hybridization analyses.
Samples were evaluated for the presence of fused or split signals in tumor cells and an estimated percentage reported. A positive result was reported when greater than 20% of tumor cells showed evidence of a rearrangement (split signal) (Figure 1). The relatively high cutoff (we usually use 5% as a cutoff) was set to allow more rigorous examination of cases falling within the 5–20% range, however, in practice this turned out to be unnecessary as all of the positive cases showed greater than 50% rearrangements. When testing was uninterpretable in a sample, it was repeated once.
Diagram of fluorescence in situ hybridization using the EWSR1 break-apart probe in extraskeletal myxoid chondrosarcoma and actual extraskeletal myxoid chondrosarcoma fluorescence in situ hybridization. Two fluorescent-labeled probes (one (G)reen and one (R)ed) hybridize the telomeric and centromeric flanking regions of EWSR1. (a) In cells negative for EWSR1 rearrangement, green and red signals are fused with the spectral overlap creating a yellow signal. (b) Cells with one green, one red signal (split signals) and one yellow (fusion) signal were considered positive for EWSR1 rearrangement.
Results
The mean age at presentation was 57-year old (range, 30–78) and patients were predominantly male (7:1). The majority of tumors (12/16) occurred in the thigh (9), inguinal (1), or buttock area (2) with the exception of 4 cases where the primary tumors occurred in the forearm, ankle, triceps, and lower leg (Table 1).
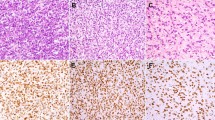
All tumors selected were at least highly suggestive of extraskeletal myxoid chondrosarcoma histologically. However, one case was much more cellular and composed of sheets/cords of epithelioid tumor cells with enlarged nuclei, coarse chromatin, and nucleoli. More characteristic features of extraskeletal myxoid chondrosarcoma were also focally present. Cases exhibiting typical histologic features and the cellular case with morphologically high-grade appearing features are illustrated in Figure 2.
Histologic examples of extraskeletal myxoid chondrosarcoma. (a) Classic extraskeletal myxoid chondrosarcoma ( × 20). Multinodular architecture with abundant myxoid material and eosinophilic cells which are more cellular in the periphery of the nodules. (b) Classic extraskeletal myxoid chondrosarcoma ( × 200). Tumor cells are spindled with eosinophilic cytoplasm and arranged in a reticular pattern. (c) Classic extraskeletal myxoid chondrosarcoma ( × 200). Epithelioid eosinophilic cells can also been seen. (d) Cellular extraskeletal myxoid chondrosarcoma with high-grade appearing features ( × 100). Highly cellular neoplasm with necrosis and hemorrhage. (e) Cellular extraskeletal myxoid chondrosarcoma with high-grade appearing features ( × 400). Tumor cells are epithelioid with enlarge nuclei, coarse chromatin, nucleoli and mitotic activity.
Fluorescence in situ hybridization was performed on 16 cases (2 metastatic tumors and 14 primary tumors) out of which 15 had sufficient analyzable interphases (Table 1). Of these 15, 14 (93%) cases were positive for EWSR1 gene rearrangement. In all cases, >50% (median 80%) of tumor nuclei were estimated to be positive for EWSR1 gene rearrangement.
Discussion
Extraskeletal myxoid chondrosarcomas harbor a balanced translocation involving the NR4A3 gene on chromosome 9 (Figure 3).8, 12 In up to 75% of cases, the NR4A3 gene is partnered in one of nine known fusion variants with the EWSR1 gene as a result of a t(9;22)(q22;q12) translocation (Figure 4).1, 2, 7, 11, 12 Other less common balanced translocations associated with extraskeletal myxoid chondrosarcoma, and involving the NR4A3 gene, include t(9;17)(q22;q11), t(9;15)(q22;q21), and t(9;3)(q22;q12).8, 19, 20 Although extraskeletal myxoid chondrosarcomas are also known to carry other recurrent and secondary chromosomal abnormalities,8 none are considered pathognomonic for this tumor, making the t(9;22) and the other rare variant translocations ideal targets for diagnostic confirmation.
Although their role is not completely understood, balanced translocations involving NRA43 are believed to be critical to the development of extraskeletal myxoid chondrosarcomas.8 The NR4A3 gene encodes an orphan nuclear receptor protein that belongs to the nuclear steroid/thyroid/retinoid receptor superfamily.8, 18 The EWSR1 gene encodes an ubiquitously expressed RNA-binding protein that may be involved in RNA transcription and unknown signal transduction pathways.8 In extraskeletal myxoid chondrosarcomas, the fusion of EWSR1–NR4A3 is thought to act as a potent transcriptional activator (aberrant transcription factor), possibly by affecting pre-mRNA splicing and resulting in the dysregulation of the unknown target genes of NR4A3.8, 18 TAF15 encodes an RNA-binding protein that shares extensive homology with proteins encoded by EWSR1 and TLS (also known as FUS), belongs to the same TET family and thus likely plays a similar biologic role in the fusion gene.8 In the other rare fusion variants, TCF12 encodes for a basic helix-loop-helix transcription factor belonging to the class A family,8, 19 whereas TFG encodes an ubiquitously expressed protein with a putative N-terminal coiled-coil domain and SPYGQ-rich regions similar to the N terminus of the TET family of proteins.8, 21 Thus both probably act in a fashion analogous to the N-terminal regions of EWSR1 and TAF15, converting NRA43 into an oncogenic fusion protein.8, 19, 20 To date, no prognostic significance of these various fusion transcripts has been described.18
Aside from conventional cytogenetic karyotyping, these translocations have been identified by other various molecular diagnostic techniques, including reverse transcription–polymerase chain reaction and fluorescence in situ hybridization (Table 2).7, 11, 12, 15, 16, 18, 22, 23
Several studies have shown reverse transcription–polymerase chain reaction to be an effective method in identifying fusion transcripts on both frozen and formalin-fixed paraffin-embedded extraskeletal myxoid chondrosarcoma tissue. The majority of these cases had t(9;22) fusion transcripts.18 Brody et al22 detected EWSR1–NRA3 fusion transcripts in six of eight (75%) tested frozen extraskeletal myxoid chondrosarcoma tissues. Similarly, Panagopoulos et al12 found an EWSR1–NR4A3 fusion variant in 15/18 (83%) tested frozen extraskeletal myxoid chondrosarcoma tissues with the remaining three cases containing a TAF15–NR4A3 fusion transcript. Sjogren et al15 also identified fusion transcripts in RNA extracted from 10 frozen tumor tissue samples from nine patients, with EWSR1–NR4A3 fusion variant accounting for 5/10 (50%) of their cases. The remaining five cases harbored a TAF15–NR4A3 (four cases including two separate metastases from the same patient) and TCF12-NR4A3 (one case) fusion transcript.15 Using reverse transcription–polymerase chain reaction on formalin-fixed paraffin-embedded tissue specimens, Antonescu et al16 found 7/9 (78%) extraskeletal myxoid chondrosarcomas to be positive for the EWSR1–NR4A3 fusion transcripts, whereas Okamoto et al11 identified EWSR1–NR4A3 (12/18, 67%) and TAF15–NR4A3 (3/18, 17%) fusion transcripts in 15/18 (83%) of their cases. Overall, these studies support the prevalence of EWSR1 gene rearrangements in extraskeletal myxoid chondrosarcoma, and reinforce the usefulness of molecular genetic testing for the differential diagnosis of extraskeletal myxoid chondrosarcomas.
A fluorescence in situ hybridization assay to detect for the rearrangement in the NRA43 locus is not commercially available. However, EWSR1 gene rearrangement fluorescence in situ hybridization analysis is available and represents an easily employed ancillary molecular diagnostic assay. In contrast to reverse transcription–polymerase chain reaction, only a few studies have examined the effectiveness of this test as an ancillary diagnostic tool for extraskeletal myxoid chondrosarcoma. Jakowski et al23 examined three cases with extraskeletal myxoid chondrosarcoma from cellblocks and found two of three had an EWSR1 gene rearranged clone by fluorescence in situ hybridization. Fluorescence in situ hybridization analysis was later repeated on the resection specimen of the negative case and was found to be positive. A more recent larger series by Downs-Kelly et al7 examined 13 extraskeletal myxoid chondrosarcomas, but found only 6/13 (46%) positive for a rearrangement of the EWSR1 by fluorescence in situ hybridization. In contrast, in our series, we found the majority of extraskeletal myxoid chondrosarcomas (14/15, 93%) to be positive for a clonal population with an EWSR1 gene rearrangement. The basis of the differences between the two studies is not entirely clear. However, it may be due to vagaries of the relatively small sample size in both studies.
Overall, most molecular studies found EWSR1 gene rearrangements present in the majority of extraskeletal myxoid chondrosarcoma cases studied (68/94, 72%), when investigated either by fluorescence in situ hybridization or reverse transcription–polymerase chain reaction. In contrast to reverse transcription–polymerase chain reaction, fluorescence in situ hybridization for EWSR1 is readily accessible due to the commercial availability of the EWSR1 probe, and its proven usefulness in the diagnosis of other neoplasms, including the Ewing family of tumors, desmoplastic small round-cell tumor, clear cell sarcoma, angiomatoid fibrous histiocytoma, and rare cases of myxoid liposarcoma (Figure 5).7, 18, 24 Rearrangements involving EWSR1 are not found in most myxoid neoplasms that can be confused with extraskeletal myxoid chondrosarcomas, including low-grade fibromyxoid sarcoma, intramuscular myxomas, chordomas, myxofibrosarcomas, and the vast majority of myxoid liposarcomas.
Major translocations in mesenchymal neoplasms involving EWSR1 and FUS (also known as TLS). EWSR1 and FUS encode for homologous proteins belonging to the same TET family. Bold lines designate the most common fusion events. EWSR1 rearrangement is not associated with the vast majority of myxoid sarcomas, which can occasionally be confused with extraskeletal myxoid chondrosarcomas.
Cellular variants of extraskeletal myxoid chondrosarcoma that mimic myxoid liposarcoma/round-cell liposarcoma and Ewing sarcoma have been reported.2, 3, 7, 11 Although the vast majority of round-cell liposarcoma have areas of more conventional myxoid liposarcoma and/or lipoblasts and most Ewing sarcomas are cytologically distinct from extraskeletal myxoid chondrosarcomas,2 distinguishing among these identities can be particularly challenging in small biopsies. Fortunately, additional molecular testing is available on paraffin-embedded formalin-fixed tissue, including fluorescence in situ hybridization to detect for the rearrangement in the CHOP/DDIT3 locus in myxoid liposarcomas and reverse transcription–polymerase chain reaction for fusion transcripts associated with Ewings family of tumors.7, 11
In conclusion, our study underscores the prevalence of EWSR1 involvement in extraskeletal myxoid chondrosarcomas. It also demonstrates that fluorescence in situ hybridization on formalin-fixed paraffin-embedded tumor tissue sections using an EWSR1 break-apart probe is a useful ancillary tool for extraskeletal myxoid chondrosarcoma. More readily available than reverse transcription–polymerase chain reaction, this assay can be useful in confirming and providing reassurance in the differential diagnosis of these rare tumors in the majority of cases.
References
Lucas DR, Heim S . Extraskeletal myxoid chondrosarcoma. In: Fletcher CDM, Unni KK, Mertens F (eds). World Health Organization of Tumors. Pathology and Genetics of Tumours of Soft Tissue and Bone, 2002 edn. IARC Press: Lyon, France, 2002, pp 213–215.
Weiss SW, Goldblum JR . Cartilaginous soft tissue tumors. In: Enzinger & Weiss's Soft Tissue Tumors, 5th edn. Mosby Elsevier: St Louis, 2008, pp 1023–1031.
Meis-Kindblom JM, Bergh P, Gunterberg B, et al Extraskeletal myxoid chondrosarcoma: a reappraisal of its morphologic spectrum and prognostic factors based on 117 cases. Am J Surg Pathol 1999;23:636–650.
Kawaguchi S, Wada T, Nagoya S, et al Extraskeletal myxoid chondrosarcoma: a multi-institutional study of 42 cases in Japan. Cancer 2003;97:1285–1292.
Oliveira AM, Sebo TJ, McGrory JE, et al Extraskeletal myxoid chondrosarcoma: a clinicopathologic, immunohistochemical, and ploidy analysis of 23 cases. Mod Pathol 2000;13:900–908.
McGrory JE, Rock MG, Nascimento AG, et al Extraskeletal myxoid chondrosarcoma. Clin Orthop Relat Res 2001;382:185–190.
Downs-Kelly E, Goldblum JR, Patel RM, et al The utility of fluorescence in situ hybridization (FISH) in the diagnosis of myxoid soft tissue neoplasms. Am J Surg Pathol 2008;32:8–13.
Hisaoka M, Hashimoto H . Extraskeletal myxoid chondrosarcoma: updated clinicopathological and molecular genetic characteristics. Pathol Int 2005;55:453–463.
Aigner T, Oliveira AM, Nascimento AG . Extraskeletal myxoid chondrosarcomas do not show a chondrocytic phenotype. Mod Pathol 2004;17:214–221.
Goh YW, Spagnolo DV, Platten M, et al Extraskeletal myxoid chondrosarcoma: a light microscopic, immunohistochemical, ultrastructural and immuno-ultrastructural study indicating neuroendocrine differentiation. Histopathology 2001;39:514–524.
Okamoto S, Hisaoka M, Ishida T, et al Extraskeletal myxoid chondrosarcoma: a clinicopathologic, immunohistochemical, and molecular analysis of 18 cases. Hum Pathol 2001;32:1116–1124.
Panagopoulos I, Mertens F, Isaksson M, et al Molecular genetic characterization of the EWS/CHN and RBP56/CHN fusion genes in extraskeletal myxoid chondrosarcoma. Genes Chromosomes Cancer 2002;35:340–352.
Sjogren H, Meis-Kindblom J, Kindblom LG, et al Fusion of the EWS-related gene TAF2N to TEC in extraskeletal myxoid chondrosarcoma. Cancer Res 1999;59:5064–5067.
Panagopoulos I, Mencinger M, Dietrich CU, et al Fusion of the RBP56 and CHN genes in extraskeletal myxoid chondrosarcomas with translocation t(9;17)(q22;q11). Oncogene 1999;18:7594–7598.
Sjogren H, Meis-Kindblom JM, Orndal C, et al Studies on the molecular pathogenesis of extraskeletal myxoid chondrosarcoma-cytogenetic, molecular genetic, and cDNA microarray analyses. Am J Pathol 2003;162:781–792.
Antonescu CR, Argani P, Erlandson RA, et al Skeletal and extraskeletal myxoid chondrosarcoma: a comparative clinicopathologic, ultrastructural, and molecular study. Cancer 1998;83:1504–1521.
Okamoto S, Hara K, Sumita S, et al Extraskeletal myxoid chondrosarcoma arising in the finger. Skeletal Radiol 2002;31:296–300.
Pfeifer JD . Tumors of soft tissue and bone. In: Molecular Genetic Testing in Surgical Pathology. Lippincott, Williams & Wilkins: Philadelphia, 2006, pp 186--231.
Sjogren H, Wedell B, Meis-Kindblom JM, et al Fusion of the NH2-terminal domain of the basic helix-loop-helix protein TCF12 to TEC in extraskeletal myxoid chondrosarcoma with translocation t(9;15)(q22;q21). Cancer Res 2000;60:6832–6835.
Hisaoka M, Ishida T, Imamura T, et al TFG is a novel fusion partner of NOR1 in extraskeletal myxoid chondrosarcoma. Genes Chromosomes Cancer 2004;40:325–328.
Hisaoka M, Okamoto S, Yokoyama K, et al Coexpression of NOR1 and SIX3 proteins in extraskeletal myxoid chondrosarcomas without detectable NR4A3 fusion genes. Cancer Genet Cytogenet 2004;152:101–107.
Brody RI, Ueda T, Hamelin A, et al Molecular analysis of the fusion of EWS to an orphan nuclear receptor gene in extraskeletal myxoid chondrosarcoma. Am J Pathol 1997;150:1049–1058.
Jakowski JD, Wakely Jr PE . Cytopathology of extraskeletal myxoid chondrosarcoma: report of 8 cases. Cancer 2007;111:298–305.
Lazar A, Abruzzo LV, Pollock RE, et al Molecular diagnosis of sarcomas: chromosomal translocations in sarcomas. Arch Pathol Lab Med 2006;130:1199–1207.
Acknowledgements
We express our appreciation to Kim-Anh Vu for her assistance in graphic design.
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclosure/conflict of interest
The authors report no conflict of interest in regards to this research.
Rights and permissions
About this article
Cite this article
Wang, WL., Mayordomo, E., Czerniak, B. et al. Fluorescence in situ hybridization is a useful ancillary diagnostic tool for extraskeletal myxoid chondrosarcoma. Mod Pathol 21, 1303–1310 (2008). https://doi.org/10.1038/modpathol.2008.114
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2008.114
Keywords
This article is cited by
-
Mutation of KIT in cellular extraskeletal myxoid chondrosarcoma: a case report and literature review
Diagnostic Pathology (2022)
-
Methylation-based classification of benign and malignant peripheral nerve sheath tumors
Acta Neuropathologica (2016)
-
Extraskeletal myxoid chondrosarcoma: a review of 23 patients treated at a single referral center with long-term follow-up
Archives of Orthopaedic and Trauma Surgery (2012)
-
Primitive neuroectodermal tumors in patients with testicular germ cell tumors usually resemble pediatric-type central nervous system embryonal neoplasms and lack chromosome 22 rearrangements
Modern Pathology (2010)