Abstract
Hypoxia-inducible factor (HIF) has important roles in promoting pro-inflammatory and bactericidal functions in myeloid cells. Conditional genetic ablation of its major subunit Hif1α in the myeloid lineage consequently results in decreased inflammatory responses in classical models of acute inflammation in mice. By contrast, we report here that mice conditionally deficient for Hif1α in myeloid cells display enhanced sensitivity to the development of airway allergy to experimental allergens and house-dust mite antigens. We support that upon allergen exposure, MyD88-dependent upregulation of Hif1α boosts the expression of the immunosuppressive cytokine interleukin (IL)-10 by lung interstitial macrophages (IMs). Hif1α-dependent IL-10 secretion is required for IMs to block allergen-induced dendritic cell activation and consequently for preventing the development of allergen-specific T-helper cell responses upon allergen exposure. Thus, this study supports that, in addition to its known pro-inflammatory activities, myeloid Hif1α possesses immunoregulatory functions implicated in the prevention of airway allergy.
Similar content being viewed by others
Introduction
Airway allergy, with allergic asthma being its most severe manifestation, is a highly prevalent chronic inflammatory disease that has reached epidemic proportions in developed countries.1 It is now well accepted that airway allergy primarily originates from the activation of dendritic cells (DCs) and the subsequent differentiation of type 2 T helper (Th2) cells in response to allergen exposure.2, 3, 4 Although the cellular and molecular mechanisms governing the development of airway allergy are starting to be well characterized, much less is known of the mechanisms that prevent the disease in healthy individuals. Yet, the identification of these mechanisms may be key to our understanding of the origins of the current airway allergy epidemic, as well as to the needed improvement of prevention strategies.1
Although a role for regulatory T cells in maintaining immune tolerance to aeroallergens has been documented in different contexts,5, 6 innate myeloid cell-mediated mechanisms seem equally important. The most documented examples to date relate to the immunoregulatory activities of lung macrophages. Two subsets of lung macrophages have been described in mice and humans, namely alveolar macrophages (AMs) and interstitial macrophages (IMs), which together represent the most abundant immune cell populations in the healthy lung.7, 8 AMs and IMs were reported to counteract the development of adaptive immune responses by inhibiting DC function.9, 10, 11 Because they are able to break the DC-mediated link between innate and adaptive immunity, AMs and IMs thus may act as first-line brakes to the development of undue immune responses to harmless inhaled antigens. Of note however, inflammatory cytokines, such as granulocyte macrophage-colony stimulating factor (GM-CSF), which are induced by common allergens and environmental Toll-like receptor (TLR) ligands,12, 13 alleviate the immunosuppressive functions of AMs.14 AMs were thus proposed to act as suppressors mainly in the absence of immunostimulatory signals in the steady state. Yet, ambient air contains a variety of immunostimulatory molecules of microbial origin, many of which are TLR ligands.15, 16, 17 IMs react to TLR activation by upregulating their production of the immunosuppressive cytokine interleukin (IL)-10, a feature absent in AMs.11 This allows IMs to inhibit DC function, and thereby the development of adaptive immune responses, in experimental models of common environmental conditions where aeroallergens are inhaled concomitantly to ambient immunostimulatory molecules.11 In spite of their potential relevance to the understanding of pulmonary immune homeostasis, the mechanisms implicated in the function of lung macrophages as myeloid regulators remain largely uncharacterized.
Hypoxia-inducible factor (HIF) is a ubiquitously expressed transcription factor mainly known for its prominent roles in regulating cellular responses to hypoxia.18 HIF is composed of the constitutively expressed Hif1β subunit (also known as ARNT) and either one of the two inducible α-subunits Hif1α and Hif2α.19 Hif1α is expressed ubiquitously, whereas expression of Hif2α is more restricted. The half-life of Hifα subunit proteins increases under hypoxia due to the inhibition of the activity of oxygen- and iron-dependent prolyl hydroxydases, which target them for proteosomal degradation under normoxic conditions.20, 21
Because of its role in the adaptation of cell metabolism to hypoxia, it was proposed that HIF is an important promoter of inflammatory responses, which require innate immune cells to migrate against oxygen gradients and to adapt to oxygen-deprived inflammatory environments.22 Indeed, mice conditionally deficient for Hif1α in myeloid cells display impaired inflammatory responses in classical models of inflammation such as forbol ester- or sodium dodecylsulfate-induced acute skin inflammation and arthritis induced by transfer of autoimmune serum.23 Myeloid HIF may also delay inflammation resolution by promoting neutrophil survival and retention at sites of inflammation.24, 25 The link between HIF and inflammation was further strengthened by the realization that HIF engages in cross-talk with another major pathway of immunity, the nuclear factor (NF)-κB pathway. Indeed, NF-κB activity upregulates Hif1α gene transcription in hypoxic conditions.26 Reciprocally, HIF may positively regulate NF-κB activity24 and NF-κB-dependent gene expression.27 Although most studies focused on hypoxic conditions, HIF promotes bactericidal and inflammatory functions of myeloid cells also in normoxia.28, 29, 30 Indeed, stimuli such as TLR ligands or inflammatory cytokines are potent inducers of HIF activity in normoxic conditions through the activation of NF-κB-dependent Hif1α transcription.28, 29, 30, 31
Here, using mouse models of conditional Hif1α deficiency, we report that, contrasting with its recognized pro-inflammatory functions, myeloid Hif1α counteracts the development of airway allergic inflammation to experimental aeroallergens and house-dust mite (HDM) antigens. We propose that myeloid Hif1α mediates these anti-inflammatory effects by positively regulating the IL-10-dependent suppressive activity of IMs on DCs upon TLR activation. Our results thus support a previously unappreciated role of HIF in maintaining mucosal immune homeostasis toward allergens in the lung.
Results
Mice deficient for Hif1α in myeloid cells display exacerbated airway allergy to HDM antigens
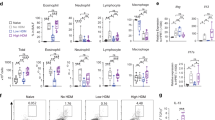
In order to study the role of Hif1α in cells of the myeloid lineage in airway allergy, we used the model of conditional genetic deletion used by Cramer et al.23 in their seminal study. This model consists in the crossing of mice with Hif1α alleles flanked by LoxP recombination sites (Hif1αfl/fl mice) with mice expressing the Cre recombinase under the dependence of the lysozyme 2 (Lysm) promoter (Lysm-Cre mice), which allows specific deletion of the Hif1α gene in cells of the myeloid lineage, namely monocytes, macrophages, and neutrophils.23 Hif1αfl/fl Lysm-Cre+ mice (Hif1αm−/− mice) were used and compared with Hif1α+/+ Lysm-Cre+ mice (Hif1α+/+ mice) as controls. Deletion of Hif1α in Hif1αm−/− mice did not significantly affect the abundance of Lysm-expressing lung myeloid cell populations, i.e. IMs, AMs, and neutrophils, or of lung DCs when compared with control mice (Figure 1a,b, and data not shown). The IM population, which is composed of CD11c− CD11b+ F4/80hi CD125− CD68+ non-autofluorescent cells, did not contain eosinophils (CD11c− CD11b+ F4/80lo CD125+ CD68− cells; data not shown). Confirming the efficiency of Cre-mediated genetic deletion in lung myeloid cells in this mouse model, real time PCR of genomic DNA from IMs and AMs isolated from the lungs of Hif1αm−/− mice indicated 80% and 90% Hif1α gene deletion, respectively (Figure 1c). Approximately 30% Hif1α gene deletion was also observed in lung DCs, which probably is explained by the monocytic origin of a fraction of lung DCs.32
Characterization of myeloid cell abundance and Hif1α gene deletion in Hif1αm−/− mice. (a) Representative plot of the flow cytometric assessment of the percentage of alveolar macrophages (AMs; F4/80+hi CD11chi autofluorescent cells), interstitial macrophages (IMs; F4/80+ CD11c− cells), and dendritic cells (DCs; F4/80−/+ CD11c+ non-autofluorescent cells) among mononucleated cells in Hif1α+/+ and Hif1αm−/− mice exposed to saline or house-dust mite (HDM) extracts for 18 h. Each plot represents an individual mouse. Similar results were obtained in six different animals, and in two independent experiments. (b) Numbers of AMs, IMs, and DCs assessed by flow cytometry as in a in the lungs of Hif1α+/+ and Hif1αm−/− mice exposed to saline or HDM extracts for 18 h. Representative data of two independent experiments with six mice/group are shown. Error bars indicate s.d. (c) Percentage of Hif1α gene deletion in the lung AMs, IMs, and DCs in Hif1αm−/− mice assessed by quantitative PCR of genomic DNA of cells purified by FACS (>95% purity). Each bar represents data obtained on DNA from indicated cell types isolated from three independent pools of lungs of 15 individuals. Data shown are representative of three independent experiments.
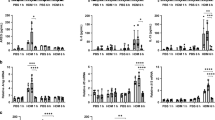
In order to compare their response to common aeroallergens, we exposed Hif1αm−/− and Hif1α+/+ mice to repeated intranasal (IN) injections of extracts of HDMs, a major source of allergens in humans. This type of regimen models physiological sensitization and elicits allergic responses that resemble human airway allergy.33, 34 We observed that Hif1αm−/− mice displayed exacerbated responses to HDM for all of the tested parameters of airway allergy. Indeed, when compared with Hif1α+/+ mice, Hif1αm−/− mice had increased inflammatory cell infiltration and mucus production in their airways (Figure 2a–c). Hif1αm−/− mice also showed increased airway resistance in response to metacholine challenge compared with control mice, as assessed using invasive measurement of dynamic resistance in mechanically ventilated mice (Figure 2d). Finally, IgE serum levels were significantly increased in Hif1αm−/− mice compared with Hif1α+/+ mice (Figure 2e). Complementary control experiments indicated that Hif1αfl/fl mice develop airway allergy similarly to Hif1α+/+ mice in this model (data not shown), thus ruling out that differences between Hif1αm−/− and Hif1α+/+ mice are due to cryptic strain-related unspecific effects.
Loss of myeloid Hif1α exacerbates airway allergy to house-dust mite (HDM) antigens in mice. Hif1α+/+ and Hif1αm−/− mice received 50 μg HDM on day 0, and 5 μg HDM on days 10 and 11, and parameters of airway allergy were assessed on day 14. (a) Total and differential immune cell counts in bronchoalveolar lavage fluid (BALF). (b) Hematoxylin and eosin and inflammatory score, and (c) quantitative real-time PCR comparison of Muc5ac mRNA expression and periodic acid-Schiff (PAS) staining and goblet cell quantification in lung sections (Bar=50 μm). (d) Measurement of dynamic airway resistance. (e) ELISA (enzyme-linked immunosorbent assay) measurement of serum immunoglobulin E (IgE) levels. Data are representative of three independent experiments with six mice/group. Error bars indicate s.d. *P<0.05, **P<0.01, ***P<0.001.
Loss of myeloid Hif1α facilitates Th2 cell priming and allergic airway sensitization
The aforementioned model suggested that Hif1αm−/− mice may be more sensitive than control mice to allergic airway sensitization. In order to test this possibility, we used a milder model of airway allergy based on intratracheal (IT) administration of the experimental antigen ovalbumin (OVA) coupled with low doses of bacterial endotoxin (lipopolysaccharide (LPS), 10 ng per treatment), followed by a short 3-day challenge with IN OVA. This model does not induce marked airway allergy in wild-type (WT) C57Bl6 mice.11 Accordingly, it primed only minor allergic responses in control Hif1α+/+ mice (Figure 3a–e). By contrast yet, Hif1αm−/− mice developed overt eosinophilic airway inflammation (Figure 3a,b), goblet cell hyperplasia (Figure 3c), airway hyperresponsiveness (Figure 3d), and increased IgE serum levels (Figure 3e) upon OVA re-exposure.
Loss of myeloid Hif1α renders mice more sensitive to allergic airway sensitization. Hif1α+/+ and Hif1αm−/− mice received 100 μg OVALPS on day 0, and were challenged intranasally with 25 μg ovalbumin (OVA) on days 12–14, and parameters of airway allergy were assessed on day 15. (a) Total and differential immune cell counts in bronchoalveolar lavage fluid (BALF). (b) Hematoxylin and eosin and inflammatory score, and (c) quantitative real-time PCR comparison of Muc5ac mRNA expression and periodic acid-Schiff (PAS) staining and goblet cell quantification in lung sections (Bar=50 μm). (d) Measurement of dynamic airway resistance. (e) ELISA (enzyme-linked immunosorbent assay) measurement of serum immunoglobulin E (IgE) levels. Data are representative of three independent experiments with five mice/group. Error bars indicate s.d. *P<0.05, **P<0.01, ***P<0.001. LPS, lipopolysaccharide; PBS, phosphate-buffered saline.
These results supported that Hif1αm−/− mice are more prone to the development of Th2 responses, the main orchestrators of airway allergy, than are control mice in response to allergen exposure. Consequently, we first assessed cell proliferation and Th2 cytokine production in the lung-draining bronchial lymph nodes (BLNs) of OVA-sensitized Hif1αm−/− and Hif1α+/+ mice upon in vitro OVA restimulation. We observed that BLN cells from Hif1αm−/− mice proliferated significantly more than cells from Hif1α+/+ mice (Figure 4a). They also produced comparatively more IL-4, IL-5, and IL-13, three type 2 cytokines (Figure 4b). Of note, we also observed increased secretion of interferon-γ (IFN-γ), which indicates that not only the Th2 component of the allergen-specific response is increased in Hif1αm−/− mice (Figure 4b). Second, we more directly studied antigen-specific T-cell proliferation by assessing the division index of adoptively transferred carboxyfluorescein succinimidyl ester (CFSE)-labeled OVA-specific OT-II cells following a single IT administration of OVA to naive Hif1αm−/−-or Hif1α+/+-recipient mice. Again, OVA-specific T cells proliferated more strongly in Hif1αm−/− mice (Figure 4c,d).
Loss of myeloid Hif1α renders mice more prone to the development of allergen-specific T-cell responses. (a) Proliferation estimated by assessment of 3H-thymidine incorporation and (b) ELISA (enzyme-linked immunosorbent assay) measurement of cytokine secretion in the supernatant of bronchial lymph node (BLN) cells isolated from Hif1α+/+ and Hif1αm−/− mice sensitized and challenged with ovalbumin (OVA) as in Figure 3 and restimulated in vitro with OVA. (c) Proliferation profile and (d) division index of adoptively transferred carboxyfluorescein succinimidyl ester (CFSE)-labeled OVA-specific CD4+ OT-II cells in the BLNs of Hif1α+/+ and Hif1αm−/− mice 3 days after intratracheal OVA instillation. (e) Proliferation estimated by assessment of 3H-thymidine incorporation and (f) ELISA measurement of cytokine secretion in the supernatant of BLN cells isolated from Hif1α+/+ and Hif1αm−/− mice sensitized and challenged with house-dust mice (HDM) as in Figure 2 and restimulated in vitro with HDM. (g) Percentages among CD4+ T cells of activated CD4+ T cells (identified as CD4+ CFSElow cells) that proliferated in response to HDM restimulation of BLN cells from Hif1α+/+ and Hif1αm−/− mice that received a single intranasal treatment with saline or HDM. (h) Representative histograms of samples compared in g. Data are representative of three independent experiments with five mice/group. Error bars indicate s.d. *P<0.05, **P<0.01, ***P<0.001. CPM, count per minute; IFN, interferon; IL, interleukin; LPS, lipopolysaccharide; PBS, phosphate-buffered saline.
Also in response to HDM exposure, Hif1αm−/− mice developed stronger Th2 responses following initial sensitization. Indeed, BLN cells from HDM-sensitized and -challenged Hif1αm−/− mice cultured in vitro in the presence of HDM proliferated significantly significantly more cells than their control counterparts (Figure 4e). They also produced more IL-4, IL-5, and IL-13, as well as IFN-γ (Figure 4f). Finally, we directly assessed HDM-specific T-cell responses in mice exposed to a single HDM treatment by isolating BLN cells, labeling them with CFSE, and restimulating them with HDM in vitro. Cell viability was high after CFSE labeling and not significantly different between BLN cells from Hif1αm−/− and Hif1α+/+ mice (data not shown). Unstimulated cells did not proliferate (data not shown). By contrast, CD4+ T cells from HDM-treated Hif1α+/+ mice proliferated in response to HDM stimulation, as evidenced by CFSE dilution, but CD4+ T cells from Hif1αm−/− mice proliferated significantly more (Figure 4g,h). Taken together, these results indicate that the absence of Hif1α in myeloid cells facilitates Th cell differentiation in response to inhaled allergens.
Hif1α in lung IMs prevents allergen-induced DC activation
The above results indicate that Hif1αm−/− mice display an overall increase in allergen-specific T-cell responses, characterized by the increased secretion of prototypical type 2 cytokines as well as IFN-γ. This may have been indicative of facilitated antigen presentation by DCs, the main triggers of allergen-specific Th cell differentiation following allergen exposure.35 We therefore compared the behavior of lung DCs in Hif1αm−/− and Hif1α+/+ mice. We observed that significantly more DCs transported antigen to the BLNs of Hif1α−/− mice after exposure to fluorescein isothiocyanate (FITC)-labeled OVA (Figure 5a,b).
Loss of myeloid Hif1α facilitates allergen-induced lung dendritic cell (DC) migration. (a) Flow cytometric assessment of the percentage and (b) numbers of FITC+ CD11c+ DCs in the BLNs of Hif1α+/+ and Hif1αm−/− mice that received OVA-FITC (ovalbumin-fluorescein isothiocyanate) intratracheally. PBS, phosphate-buffered saline.
This observation suggested that a brake to DC activation by allergens was lost following deletion of myeloid Hif1α. We therefore studied the effect of myeloid Hif1α deletion on the function of IMs and AMs, which were previously shown to counteract DC activation following antigen exposure.10, 11 To compare the immunosuppressive function of control and Hif1α−/− IMs on DCs, we isolated IMs from Hif1αm−/− and Hif1α+/+ mice and cultured them with syngeneic WT bone marrow–derived DCs (BMDCs).
Overnight pulse with OVA induces the maturation of BMDCs, as indicated by their increased levels of cell-surface expression of MHC (major histocompatibility complex) class II and CD86 (Figure 6a). Co-culture with Hif1α+/+ IMs significantly impaired OVA-induced upregulation of MHC II and CD86 in BMDCs, whereas Hif1α−/− IMs failed to display similar activity on BMDC maturation (Figure 6a).
Hif1α-deficient lung interstitial macrophages (IMs) have impaired immunosuppressive activity on dendritic cells (DCs). (a) Flow cytometric assessment of the expression levels of MHC-II (major histocompatibility complex-II) and CD86 at the surface of bone marrow–derived DCs (BMDCs) cultured alone (OVALPS-BMDCs) or with syngeneic IMs isolated from Hif1α+/+ (OVALPS-BMDCs/Hif1α+/+ IMs) or Hif1αm−/− mice (OVALPS-BMDCs/Hif1α−/− IMs). (b–g) Airway allergy parameters in ovalbumin (OVA)–challenged wild-type mice that received OVALPS-BMDCs, OVALPS-BMDCs/Hif1α+/+ IMs, or OVALPS-BMDCs/Hif1α−/− IMs. (b) Total and differential immune cell counts in bronchoalveolar lavage fluid (BALF). (c) Hematoxylin and eosin and inflammatory score, and (d) quantitative real-time PCR comparison of Muc5ac mRNA expression and periodic acid-Schiff (PAS) staining and goblet cell quantification in lung sections (Bar=50 μm). (e) ELISA (enzyme-linked immunosorbent assay) measurement of serum immunoglobulin E (IgE) levels. (f) Proliferation estimated by assessment of 3H-thymidine incorporation and (g) ELISA measurement of cytokine secretion in the supernatant of in vitro OVA-restimulated bronchial lymph node cells. Data are representative of two (a) or three (b–g) independent experiments with three (a) or five (b–g) mice/group. Error bars indicate s.d. *P<0.05, **P<0.01, ***P<0.001. CPM, count per minute; IFN, interferon; IL, interleukin; LPS, lipopolysaccharide; MFI, mean fluorescence intensity; PBS, phosphate-buffered saline.
We transferred OVA-pulsed co-cultures of IMs and BMDCs IT to naive recipient WT mice. Upon airway challenge with aerosolized OVA, mice that received OVA-pulsed WT BMDCs alone developed marked airway inflammation (Figure 6b,c), goblet cell hyperplasia (Figure 6d), and elevated serum IgE levels (Figure 6e). In agreement with previous findings,11 these responses were inhibited by the culture of BMDCs in the presence of control IMs (Figure 6b–e). By contrast, Hif1α−/− IMs were unable to prevent BMDC-induced airway allergy in this model (Figure 6b–e). Accordingly, BLN cell proliferation and Th2 cytokine secretion were comparable in mice that received BMDCs co-cultured with Hif1α−/− IMs and mice that received BMDCs alone, whereas they were significantly inhibited in mice that received BMDCs co-cultured with control IMs (Figure 6f,g).
These results thus indicate that Hif1α-deficient IMs have impaired immunoregulatory activities upon allergen exposure. Similar experiments performed with Hif1α+/+ and Hif1α−/− AMs in place of IMs indicated that neither Hif1α+/+ nor Hif1α−/− AMs are able to interfere with DC function in this model (data not shown), in agreement with previous findings using WT AMs.11
MyD88-dependent activation of Hif1α boosts the immunosuppressive activity of IMs
We aimed to determine how allergens may upregulate Hif1α activity in IMs, and how Hif1α may in turn impact on IM regulatory activity. HDM preparations notably contain LPS, and the major dust mite allergen Der p 2 was shown to mimic the structure and activity of the MD2 subunit of the endotoxin receptor complex.36 We thus suspected that, as shown in other experimental contexts,29 TLR engagement by HDM may promote Hif1α gene transcription in IMs. We indeed observed that HDM-induced upregulation of Hif1α mRNA expression was abrogated in MyD88-deficient IMs (Figure 7a).
Allergens boost interleukin (IL)-10 production by lung IMs through MyD88-dependent signaling. (a) Quantitative PCR comparison of Hif1α mRNA expression in interstitial macrophages (IMs) isolated from the lung of wild-typeand MyD88α−/− mice and stimulated with house-dust mite (HDM) for 4 h. (b) ELISA (enzyme-linked immunosorbent assay) measurement of IL-10 and transforming growth factor (TGF)-β production by IMs isolated from the lung of Hif1α+/+ and Hif1αm−/− mice and stimulated with HDM for 18 h. Error bars indicate s.d. *P<0.05. Data are representative of three independent experiments. PBS, phosphate-buffered saline.
IMs were previously shown to inhibit DC function by their upregulation of IL-10 secretion in response to TLR activation.11 We observed that Hif1α−/− IMs secreted significantly less IL-10 than their control counterparts in response to HDM, while their production of transforming growth factor-β (TGF-β) was comparable (Figure 7b).
We consequently further tested the role of IL-10 in the regulatory activity of IMs, and whether impaired secretion of IL-10 accounts for the deficient activity of Hif1α−/− IMs. In coculture experiments in which IMs were separated of BMDCs by a semipermeable membrane, we neutralized IL-10 secreted by Hif1α+/+ IMs using monoclonal antibodies. Symmetrically in a similar setting, we complemented the supernatant of co-cultures of BMDCs and Hif1α−/− IMs with recombinant mouse IL-10 to reach concentrations similar to those observed with Hif1α+/+ IMs. The BMDCs were subsequently separately transferred to naive recipient mice and their ability to induce airway allergic sensitization was assessed. We observed that transfer of BMDCs co-cultured with Hif1α+/+ IMs and neutralizing anti-IL-10 antibodies led to increased parameters of airway allergy (Figure 8a–c) and allergen-specific BLN cell proliferation and type 2 cytokine and IFN-γ secretion (Figure 8d,e) following allergen re-exposure, as compared with mice that received BMDCs co-cultured with Hif1α+/+ IMs and isotype control antibodies. Of note, the measured parameters in these mice were similar to those of mice that received BMDCs co-cultured with Hif1α−/− IMs. Reciprocally, mice that received BMDCs co-cultured with IL-10-complemented Hif1α−/− IMs displayed reduced parameters of airway allergy (Figure 8a–c) and allergen-specific BLN cell proliferation and type 2 cytokine and IFN-γ secretion (Figure 8d,e) following allergen re-exposure compared with mice that received BMDCs co-cultured with excipient-treated Hif1α−/− IMs. Parameters of airway allergy in these mice resembled those of mice that received BMDCs co-cultured with Hif1α+/+ IMs.
Impaired secretion of interleukin (IL)-10 accounts for reduced immunoregulatory activity of Hif1α−/− interstitial macrophages (IMs). (a–g) Airway allergy parameters in ovalbumin (OVA)-challenged wild-type mice that received bone marrow–derived dendritic cells (BMDCs) cultured alone (OVALPS-BMDCs) or with syngeneic IMs isolated from Hif1α+/+ (OVALPS-BMDCs/Hif1α+/+ IMs) or Hif1αm−/− (OVALPS-BMDCs/Hif1αm−/− IMs) mice. Some co-cultures of BMDCs with Hif1α+/+ IMs were treated with neutralizing anti-IL-10 antibodies (OVALPS-BMDCs/Hif1α+/+ IMs + αIL-10), whereas some co-cultures of BMDCs with Hif1α−/− IMs were treated with recombinant mouse IL-10 (OVALPS-BMDCs/Hif1α−/− IMs+rIL-10). To allow comparison between groups, isotype-control antibodies (Iso) were added to all appropriate co-cultures. (a) Total and differential immune cell counts in bronchoalveolar lavage fluid (BALF). (b) Hematoxylin and eosin and inflammatory score, and (c) quantitative real-time PCR comparison of Muc5ac mRNA expression and periodic acid-Schiff (PAS) staining and goblet cell quantification in lung sections (Bar=50 μm). (d) Proliferation estimated by assessment of 3H-thymidine incorporation and (e) ELISA (enzyme-linked immunosorbent assay) measurement of cytokine secretion in the supernatant of in vitro OVA-restimulated bronchial lymph node cells. Data are representative of two (a) or three (b–e) independent experiments with three (a) or five (b–e) mice/group. Error bars indicate s.d. *P<0.05, **P<0.01, ***P<0.001. CPM, count per minute; IFN, interferon; LPS, lipopolysaccharide; PBS, phosphate-buffered saline.
Finally, to directly assess whether impaired IL-10 secretion accounts for the allergy-prone phenotype of Hif1αm−/− mice, we assessed the effects of impacting on IL-10 signaling in allergen-exposed Hif1α+/+ and Hif1αm−/− mice. We observed that administration of neutralizing anti-IL-10 antibodies at the time of allergen exposure facilitated airway allergic sensitization in Hif1α+/+ mice (Figure 9a–c) as well as allergen-specific BLN cell proliferation and type 2 cytokine and IFN-γ secretion (Figure 9d,e), phenocopying the situation in Hif1αm−/− mice. Symmetrically, local administration of recombinant murine IL-10 counteracted airway allergic sensitization in Hif1αm−/− mice (Figure 9a–c) and decreased allergen-specific BLN cell proliferation and type 2 cytokine and IFN-γ secretion (Figure 9d,e), mimicking the situation in Hif1α+/+ mice. Secretion of IL-10 and TGF-β was similar in allergen-challenged BLN cells from Hif1α+/+ and Hif1αm−/− mice and was not affected by the administration of neutralizing anti-IL-10 antibodies or recombinant IL-10 (Figure 9f), suggesting that myeloid Hif1α does not impact on regulatory T-cell responses.
Impaired interleukin (IL)-10 signaling accounts for the increased susceptibility of Hif1αm−/− mice to airway allergic sensitization. (a–g) Airway allergy parameters in Hif1α+/+ and Hif1αm−/− mice sensitized and challenged with ovalbumin (OVA) as in Figure 3. A group of Hif1α+/+ mice was treated with neutralizing anti-IL-10 antibodies (αIL-10) and a group of Hif1αm−/− mice received recombinant IL-10 (rIL-10) at the time of allergenic sensitization. To allow comparisons between groups, all appropriate mouse groups received isotype-control antibodies (Iso). (a) Total and differential immune cell counts in bronchoalveolar lavage fluid (BALF). (b) Hematoxylin and eosin and inflammatory score, and (c) quantitative real-time PCR comparison of Muc5ac mRNA expression and periodic acid-Schiff (PAS)-staining and goblet cell quantification in lung sections (Bar=50 μm). (d) Proliferation estimated by the assessment of 3H-thymidine incorporation and (e, f) ELISA (enzyme-linked immunosorbent assay) measurement of cytokine secretion in the supernatant of in vitro OVA-restimulated bronchial lymph node cells. Data are representative of two (a) or three (b–f) independent experiments with three (a) or five (b–f) mice/group. Error bars indicate s.d. *P<0.05, **P<0.01, ***P<0.001. CPM, count per minute; IFN, interferon; LPS, lipopolysaccharide; PBS, phosphate-buffered saline; TGF, transforming growth factor.
Altogether, these results support that Hif1α controls the immunoregulatory activity of IMs toward DCs by upregulating their production of IL-10 in response to TLR activation and that Hif1-dependent IL-10 secretion in turns counteracts airway allergic sensitization.
Discussion
Airway allergy results from the development of aberrant adaptive responses against common environmental aeroantigens. We show here that deletion of Hif1α in myeloid cells unexpectedly results in increased sensitivity to airway allergy in mice. This observation supports the existence of previously unappreciated Hif-1-dependent protective mechanisms responsible for maintaining immunological homeostasis toward allergens at lung mucosal surfaces.
So far, known mechanisms of Hif1α-dependent protection of mucosal surfaces mainly involved Hif1α as a promoter of the bactericidal and inflammatory function of myeloid cells, actively protecting mucosal surfaces from pathogens.22 Moreover, Hif1α was recently identified as an orchestrator of the differentiation of T cells into Th17 cells and as a negative regulator of regulatory T-cell differentiation.37, 38 By contrast with these pro-inflammatory functions of Hif1α in immune cells, epithelial Hif1α may promote the expression of anti-inflammatory mediators such as netrin-1 or key actors of adenosine metabolism and signaling, which downregulate immune cell function.22, 39, 40 Epithelial Hif1α would in this way keep the pro-inflammatory functions of immune cells in check and protect mucosal surfaces from inflammation-induced injury.
Hif1α-dependent protection of mucosal surfaces thus so far seemed to depend on the compartmentalization of pro- and anti-inflammatory activities of Hif1α in immune and epithelial cells, respectively. Our results contrast with this clear-cut division of Hif1α function. Myeloid Hif1α could also indeed act as an early checkpoint for the development of adaptive responses against aeroantigens and thereby protect respiratory mucosal surfaces from undue inflammatory responses. In consequence, this study supports that Hif1α has an ambivalent role in immune cells, acting as a pro-inflammatory or anti-inflammatory transcription factor in a context- and cell-dependent manner in physiological conditions. This proposal may be related to the recent suggestion that, in pathological conditions, Hif1α may impair tumor immune surveillance by promoting immunosuppressive activities in myeloid suppressor cells.41, 42
Previous studies using widespread gene deletion or pharmacological inhibition in mice supported a neat pro-inflammatory function of HIF in airway allergy.43, 44 Although the cellular and molecular mechanisms underlying these observations were not investigated in detail, the anti-inflammatory outcome of general HIF inhibition most likely is linked to the requirement for HIF in the maturation and function of DCs.45, 46, 47 Again, the fact that the outcome of HIF impairment depends on the cell types targeted highlights the importance of cellular compartmentalization in determining HIF function.
Our results support that facilitation of airway allergy development upon deletion of myeloid Hif1α occurs primarily through the loss of the immunoregulatory activity of IMs. IMs were recently proposed as essential regulators of pulmonary immune homeostasis toward aeroantigens,11 although the mechanisms governing their function remain to be established. We observed here that IMs require intact Hif1α activity to optimally secrete IL-10 and antagonize DC activation following allergen stimulation. Hif1α activity in IMs would thereby protect respiratory mucosal surfaces from undue DC-induced adaptive immune responses. Supporting this notion, Hif1αm−/− mice and mice depleted of their IMs using depleting antibodies display comparably increased susceptibility to the development of experimental airway allergy.11
In addition to IMs, the model of conditional ablation used in this study also induced Hif1α gene deletion in AMs, neutrophils, and a fraction of lung DCs. AMs have been reported to possess immunosuppressive activities in certain models of antigen exposure.9, 10 However, in agreement with a previous report, we did not observe any inhibitory effect of AMs on DC function upon OVALPS exposure in vitro.11 Furthermore, using IT administration of clodronate-loaded liposomes to deplete AMs as in previous studies,9, 10 it was shown that AMs are not able to interfere with allergic airway sensitization in the model of OVA-induced airway allergy used in the present study.11 Similarly, we observed that AM depletion did not impact on the development of airway allergy in our model of HDM exposure (M. Toussaint, unpublished observations). Altogether, these observations reduce the likeliness of a role for AMs in the prevention of airway allergic sensitization through Hif1α-dependent immunoregulatory activities. We nevertheless do not rule out that AMs may impact on parameters of airway allergy in sensitized subjects through other mechanisms, as has been proposed by previous studies.48, 49 The role of neutrophils in the pathogenesis of airway allergy is still unclear. Although potential immunoregulatory effects of certain neutrophil products have been proposed by a few in vitro studies,50, 51, 52 most experimental and clinical evidence tends to link neutrophils with pro-inflammatory activities and aggravated asthmatic phenotypes.53, 54, 55 Although we do not formally exclude this hypothesis, it is unlikely that Hif1α in neutrophils could counteract airway allergy. Finally, Hif1α has been shown to promote the pro-inflammatory and pro-Th2 functions of DCs.45, 46, 47 It is thus unlikely that Hif1α deletion in a fraction of lung DCs would result in anti-inflammatory activities. We thus support that, even though we do not formally exclude a putative participation of other cell types, IMs are the primary cells in which myeloid Hif1α has its immunoregulatory role in response to aeroallergens.
IMs share numerous phenotypic and functional similarities with macrophages of the intestinal lamina propria (LPMs). Indeed, both IM and LPMs share similarities in their cell-surface phenotype and react to TLR ligands by upregulating their production of IL-10, which in turn contributes to preventing the development of undue adaptive responses, respectively, in the lung11 and intestine.56 Although the concept is only emerging and will require further investigation, it is tempting to consider IMs and LPMs as a unique type of regulatory macrophages, in charge of ensuring immune homeostasis at mucosal sites permanently exposed to external stimuli. Even though further experiments are needed to confirm this hypothesis in LPMs, we propose that HIF activity may be a central determinant of the immunoregulatory function of regulatory macrophages at mucosal surfaces.
Methods
Mice. Hif1αfl/fl,57 Lysm-Cre+58 and OVA-specific, MHC II–restricted, T-cell receptor (TCR)–transgenic OT-II (H-2b) mice were purchased from the Jackson Laboratory (Bar Harbor, ME). All transgenic mice were on a C57Bl/6 background. All mice were bred and housed in institutional specific pathogen-free facilities. Age-paired groups of females 8–10 weeks of age were used. All experimental procedures and protocols were approved by the Institutional Animal Care and Use Committee of the University of Liege. We also followed the “Guide for the Care and Use of Laboratory Animals” prepared by the Institute of Laboratory Animal Resources, National Research Council and published by the National Academy Press (revised 1996).
Reagents. Lyophilized HDM (Dermatophagoides pteronyssinus) extract was from Greer Labs (Lenoir, NC; endotoxin contents: 0.84 ng mg−1, i.e. 0.084 ng per IN administration). LPS-free OVA (EndoGrade Ovalbumin; endotoxin concentration <0.1 ng mg−1) was from Hyglos (Bernried am Starnberger See, Germany). Chicken OVA, LPS from Escherichia coli (serotype 055:B5), and methacholine were purchased from Sigma-Aldrich (St Louis, MO). LPS-free OVA-FITC was provided by Dr Murielle Pichavant (Institut Pasteur de Lille, France). Recombinant murine GM-CSF was provided by Dr Kris Thielemans (Medical School of the Vrije Universiteit Brussel, Belgium). 7-Aminoactnomycin (7-AAD) was from BD Pharmingen (San Diego, CA). Recombinant mouse IL-10 was from eBiosciences (San Diego, CA).
Antibodies. V450-conjugated anti-CD11b (M1/70) and Alexa Fluor 488–conjugated anti-CD125 (T21) antibodies were from BD Biosciences (San Jose, CA). Allophycocyanin-eFluor-conjugated anti-CD11c (N418), allophycocyanin-conjugated anti-TCR Vα2 (B20.1), Pacific Blue–conjugated anti-CD4 (RM4-5), phycoerythrin-streptavidin and biotinylated anti-F4/80 (BM8) and neutralizing monoclonal anti-IL-10 antibodies were from eBioscience. FITC-conjugated anti-CD68 (FA-11) was from AbD Serotec (Raleigh, NC). 2.4G2 Fc receptor antibodies were produced in house.
Cell sorting and flow cytometry. To obtain single-lung-cell suspensions, lungs were perfused with phosphate-buffered saline (PBS) through the right ventricle, cut into small pieces, and digested for 1 h at 37°C in 1 mg ml−1 collagenase A (Roche Diagnostics, Indianapolis, IN) and 0.05 mg ml−1 DNaseI (Roche Diagnostics) in Hank’s balanced salt solution. IMs, AMs, and DCs were sorted by flow cytometry on a FACSAria (BD Biosciences) based on their differential expression of F4/80, CD11c, and CD11b as previously described.11 Flow cytometry analyses were performed on a FACScanto II (BD Biosciences). Staining reactions were performed at 4 °C. Cells were incubated with 2.4G2 Fc receptor antibodies to reduce non-specific binding. Cell viability was assessed using 7-AAD.
Determination of allele-deletion frequency by quantitative PCR. Genomic DNA from IMs, AMs, and DCs sorted from the lung of Hif1α+/+ and Hif1αm−/− mice was isolated using High Pure PCR Template Preparation Kits (Roche Diagnostics). The following primers were used: Hif1α-forward 5′-TGAGCTTGCTCATCAGTTGC-3′ and Hif1α-reverse 5′-GGTGAGCCTCATAACAGAAGC-3′. Real-time PCR analyses on 20 ng DNA were performed using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) and the iQ5 real-time PCR detection system (Bio-Rad). The degree of excision was calculated by comparison of Hif1α-intact DNA relative to an unexcised gene.
Models of airway allergy. In models of HDM-induced airway allergy, lightly isoflurane-anesthetized mice received an IN instillation of vehicle (LPS-free saline) or HDM extract on day 0 (50 μg in 50 μl) and on days 10 and 11 (5 μg in 50 μl). Mice were killed 3 days after the last HDM treatment. In models of OVA-induced airway allergy, isoflurane-anesthetized mice received an IT administration of LPS-free OVA combined with a low dose of LPS (OVALPS; OVA: 100 μg per mouse; LPS: 10ng per mouse, in 50 μl PBS). From days 12 to 14, mice were challenged IN with 25 μg OVA (grade V; Sigma-Aldrich) in 50 μl PBS. Mice were killed on day 15. In some experiments, mice received 200 μg neutralizing anti-IL-10 antibodies intravenously 90 min before and 1 h after IT sensitization with OVALPS. Alternatively, mice received 3 μg recombinant mouse IL-10 mixed with the sensitizing dose of OVALPS. Parameters of allergy in these models, including cytology of the bronchoalveolar lavage fluid, lung histology, and total IgE serum levels, were assessed as described previously.11, 34 The extent of peribronchial inflammation was estimated by a score calculated by quantification of peribronchial inflammatory cells in lung sections stained with hematoxylin and eosin, as described previously.11 Mucus production was quantified as percentage of periodic acid-Schiff-stained goblet cells per total epithelial cells in randomly selected bronchi. Seven randomly selected sections were analyzed per mouse lung. Airway hyper-responsiveness was evaluated by measuring dynamic airway resistance in anesthetized mice using a flexiVent small animal ventilator (SCIREQ, Tempe, AZ), as previously described.11 Changes were expressed as percentage change from the baseline measurement (performed following PBS exposure).
Restimulation of BLN cells. BLN cells (2 × 105 cells in a 96-well plate) were cultured in Click's medium supplemented with 0.5% heat-inactivated mouse serum and additives, with or without HDM (30 μg ml−1) or OVALPS (50 μg ml−1). The proliferation was measured as 3H-thymidine incorporation during the last 16 h of a 4-day culture. Culture supernatants were assayed for IL-4, IL-5, IL-10, IL-13, IFN-γ, and TGF-β by ELISA (enzyme-linked immunosorbent assay; Biosource/Invitrogen, Carlsbad, CA).
Assessment of allergen-specific CD4+ T-cell activation. In certain models, on day −1, splenic cells from OT-II mice (5 × 107 cells ml−1) were labeled with CFSE (0.5 μm in PBS) for 10 min at 37 °C and washed. In all, 10 × 106 cells were then administered in the tail vein of recipient mice. On day 0, mice were injected IT with vehicle or 100 μg OVALPS in 50 μl PBS. On day 3, BLN T-cell responses were analyzed by assessing CFSE dilution in TCR Vα2+ CD4+ T cells. Alternatively, mice were administered 50 μg of HDM or vehicle IN. Five days later, BLN cells were labeled with CFSE and cultured in vitro for 5 days in complete Click's medium supplemented with 0.5% heat-inactivated mouse serum and additives, with or without HDM (30 μg ml−1). Cells were stained for CD4 and analyzed for CFSE dilution by flow cytometry. Division index was calculated using Flow-Jo software (Tree Star, Ashland, OR) to estimate T-cell proliferation.
DC migration assay. The antigen uptake and migratory activity of lung DCs were evaluated by IT administration of 100 μg FITC-labeled OVA combined with 10 ng LPS, followed 18 h later by flow cytometric analysis of the numbers of F4/80− CD11c+ FITC+ DCs in the BLNs.
Cocultures. BMDCs were generated as described previously.11 At day 8, the culture medium was replaced with medium devoid of GM-CSF. FACS-sorted IMs were added to BMDC cultures at a ratio of 1:1, 1 h before treatment with OVALPS or PBS. In some experiments, BMDCs and IMs were co-cultured separated by a semi-permeable membrane (1-μm pore culture inserts; BD Biosciences) with 10 μg ml−1 neutralizing anti-IL-10 antibodies or 1 ng ml−1 recombinant mouse IL-10. On day 9, 1 × 106 BMDCs, alone or with the co-cultured IMs, were injected IT to recipient mice and 10 days later, mice were challenged with OVA (1% wt/vol in PBS; grade III; Sigma-Aldrich) aerosol during a daily 30-min challenge on 5 consecutive days. Viability of IMs and BMDCs before adoptive transfer was assessed by 7-AAD staining and flow cytometric analysis. Twenty-four hours after the last challenge, AHR was measured, the mice were killed, and airway allergy was evaluated.
Production of IL-10 and TGF-β by IMs. FACS-sorted IMs were stimulated with HDM (30 μg ml−1) or excipient and cultivated for 16 h. Culture supernatants were assayed for IL-10 and TGF-β by ELISA (eBioscience).
Quantitative PCR analysis of Hif1α and Muc5ac mRNA. For the analysis of Hif1α mRNA, the RNA of FACS-sorted IMs from mice exposed to 50 μg ml−1 HDM for 4 h was isolated using TRIzol (Invitrogen). For the analysis of Muc5ac mRNA expression, mouse lungs were crushed and homogenized in TRIzol, and RNA was extracted. One microgram of RNA by sample was used for reverse transcription reactions using the RevertAid First Strand cDNA Synthesis Kit (Fermentas, Waltham, MA). Specific primers with the following sequences were used: Hif1α-forward 5′-TAAACCTGGCAATGTCTCCTT-3′, Hif1α-reverse 5′-TCAGTGCAGGATCAGCACTAC-3′, Muc5ac-forward: 5′-TACAATGGGCAACGGTACCATCCT-3′, Muc5ac-reverse: 5′-AACTGCAGGTGTCAACGATCCTCT-3′ and for the references genes: Actb-forward 5′-AGGTGACAGCATTGCTTCTG-3′, Actb-reverse 5′-GCTGCCTCAACACCTCAAC-3′; Gapdh-forward 5′-TGGCAAAGTGGAGATTGTTGCC-3′, Gapdh-reverse 5′-AAGATGGTGATGGGCTTCCCG-3′; Hprt-forward 5′-AGCTACTGTAATGATCAGTCAACG-3′, and Hprt-reverse 5′-AGAGGTCCTTTTCACCAGCA-3′. Quantification and normalization of the results were performed using the qBase+ software (Biogazelle, Ghent, Belgium). All samples were run in triplicate for the target and reference genes.
Statistical analysis. Data are presented as means±standard deviation (s.d.). The differences between mean values were estimated using pairwise Student's t-tests following Anderson–Darling tests for assessment of normality of the data distributions. In all the figures, asterisks are used to indicate significant differences observed when comparing Hif1α+/+- and Hif1αm−/−-treated groups (*P<0.05, **P<0.01, ***P<0.001).
References
Eder, W., Ege, M.J. & von Mutius, E. The asthma epidemic. N. Engl. J. Med. 355, 2226–2235 (2006).
Barrett, N.A. & Austen, K.F. Innate cells and T helper 2 cell immunity in airway inflammation. Immunity 31, 425–437 (2009).
Lambrecht, B.N. & Hammad, H. Biology of lung dendritic cells at the origin of asthma. Immunity 31, 412–424 (2009).
Locksley, R.M. Asthma and allergic inflammation. Cell 140, 777–783 (2010).
Robinson, D.S. Regulatory T cells and asthma. Clin. Exp. Allergy 39, 1314–1323 (2009).
Arnold, I.C. et al. Helicobacter pylori infection prevents allergic asthma in mouse models through the induction of regulatory T cells. J. Clin. Invest. 121, 3088–3093 (2011).
Franke-Ullmann, G., Pfortner, C., Walter, P., Steinmuller, C., Lohmann-Matthes, M.L. & Kobzik, L. Characterization of murine lung interstitial macrophages in comparison with alveolar macrophages in vitro. J. Immunol. 157, 3097–3104 (1996).
Hoppstadter, J. et al. Differential cell reaction upon Toll-like receptor 4 and 9 activation in human alveolar and lung interstitial macrophages. Respir. Res. 11, 124 (2010).
Thepen, T., Van Rooijen, N. & Kraal, G. Alveolar macrophage elimination in vivo is associated with an increase in pulmonary immune response in mice. J. Exp. Med. 170, 499–509 (1989).
Holt, P.G. et al. Downregulation of the antigen presenting cell function(s) of pulmonary dendritic cells in vivo by resident alveolar macrophages. J. Exp. Med. 177, 397–407 (1993).
Bedoret, D. et al. Lung interstitial macrophages alter dendritic cell functions to prevent airway allergy in mice. J. Clin. Invest. 119, 3723–3738 (2009).
King, C., Brennan, S., Thompson, P.J. & Stewart, G.A. Dust mite proteolytic allergens induce cytokine release from cultured airway epithelium. J. Immunol. 161, 3645–3651 (1998).
Hammad, H., Chieppa, M., Perros, F., Willart, M.A., Germain, R.N. & Lambrecht, B.N. House dust mite allergen induces asthma via Toll-like receptor 4 triggering of airway structural cells. Nat. Med. 15, 410–416 (2009).
Bilyk, N. & Holt, P.G. Inhibition of the immunosuppressive activity of resident pulmonary alveolar macrophages by granulocyte/macrophage colony-stimulating factor. J. Exp. Med. 177, 1773–1777 (1993).
Michel, O. et al. Severity of asthma is related to endotoxin in house dust. Am. J. Respir. Crit. Care Med. 154 (6 part 1), 1641–1646 (1996).
Thorn, J. & Rylander, R. Airways inflammation and glucan in a rowhouse area. Am. J. Respir. Crit. Care Med. 157, 1798–1803 (1998).
Douwes, J. et al. Does early indoor microbial exposure reduce the risk of asthma? The Prevention and Incidence of Asthma and Mite Allergy Birth Cohort Study. J. Allergy Clin. Immunol. 117, 1067–1073 (2006).
Weidemann, A. & Johnson, R.S. Biology of HIF-1alpha. Cell. Death Differ. 15, 621–627 (2008).
Wang, G.L., Jiang, B.H., Rue, E.A. & Semenza, G.L. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA 92, 5510–5514 (1995).
Bruick, R.K. & McKnight, S.L. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294, 1337–1340 (2001).
Jaakkola, P. et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472 (2001).
Sitkovsky, M. & Lukashev, D. Regulation of immune cells by local-tissue oxygen tension: HIF1 alpha and adenosine receptors. Nat. Rev. Immunol. 5, 712–721 (2005).
Cramer, T. et al. HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell 112, 645–657 (2003).
Walmsley, S.R. et al. Hypoxia-induced neutrophil survival is mediated by HIF-1alpha-dependent NF-kappaB activity. J. Exp. Med. 201, 105–115 (2005).
Elks, P.M. et al. Activation of hypoxia-inducible factor-1alpha (Hif-1alpha) delays inflammation resolution by reducing neutrophil apoptosis and reverse migration in a zebrafish inflammation model. Blood 118, 712–722 (2011).
Rius, J. et al. NF-kappaB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1alpha. Nature 453, 807–811 (2008).
Peyssonnaux, C., Cejudo-Martin, P., Doedens, A., Zinkernagel, A.S., Johnson, R.S. & Nizet, V. Cutting edge: essential role of hypoxia inducible factor-1alpha in development of lipopolysaccharide-induced sepsis. J. Immunol. 178, 7516–7519 (2007).
Peyssonnaux, C. et al. HIF-1alpha expression regulates the bactericidal capacity of phagocytes. J. Clin. Invest. 115, 1806–1815 (2005).
Jantsch, J. et al. Toll-like receptor activation and hypoxia use distinct signaling pathways to stabilize hypoxia-inducible factor 1alpha (HIF1A) and result in differential HIF1A-dependent gene expression. J. Leukoc. Biol. 90, 551–562 (2011).
Mahabeleshwar, GH. et al. The myeloid transcription factor KLF2 regulates the host response to polymicrobial infection and endotoxic shock. Immunity 34, 715–728 (2011).
Blouin, C.C., Page, E.L., Soucy, G.M. & Richard, D.E. Hypoxic gene activation by lipopolysaccharide in macrophages: implication of hypoxia-inducible factor 1alpha. Blood 103, 1124–1130 (2004).
Landsman, L., Varol, C. & Jung, S. Distinct differentiation potential of blood monocyte subsets in the lung. J. Immunol. 178, 2000–2007 (2007).
Cates, E.C. et al. Intranasal exposure of mice to house dust mite elicits allergic airway inflammation via a GM-CSF-mediated mechanism. J. Immunol. 173, 6384–6392 (2004).
Marichal, T. et al. Interferon response factor 3 is essential for house dust mite-induced airway allergy. J. Allergy Clin. Immunol. 126, 836–844 (2010).
Hammad, H. et al. Inflammatory dendritic cells—not basophils—are necessary and sufficient for induction of Th2 immunity to inhaled house dust mite allergen. J. Exp. Med. 207, 2097–2111 (2010).
Trompette, A. et al. Allergenicity resulting from functional mimicry of a Toll-like receptor complex protein. Nature 457, 585–588 (2009).
Shi, LZ. et al. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J. Exp. Med. 208, 1367–1376 (2011).
Dang, E.V. et al. Control of TH17/Treg balance by hypoxia-inducible factor 1. Cell 146, 772–784 (2011).
Thiel, M. et al. Oxygenation inhibits the physiological tissue-protecting mechanism and thereby exacerbates acute inflammatory lung injury. PLoS. Biol. 3, e174 (2005).
Rosenberger, P. et al. Hypoxia-inducible factor-dependent induction of netrin-1 dampens inflammation caused by hypoxia. Nat. Immunol. 10, 195–202 (2009).
Corzo, CA. et al. HIF-1alpha regulates function and differentiation of myeloid-derived suppressor cells in the tumor microenvironment. J. Exp. Med. 207, 2439–2453 (2010).
Doedens, A.L. et al. Macrophage expression of hypoxia-inducible factor-1 alpha suppresses T-cell function and promotes tumor progression. Cancer Res. 70, 7465–7475 (2010).
Huerta-Yepez, S. et al. Hypoxia inducible factor promotes murine allergic airway inflammation and is increased in asthma and rhinitis. Allergy 66, 909–918 (2011).
Kim, S.R. et al. HIF-1alpha inhibition ameliorates an allergic airway disease via VEGF suppression in bronchial epithelium. Eur. J. Immunol. 40, 2858–2869 (2010).
Jantsch, J. et al. Hypoxia and hypoxia-inducible factor-1alpha modulate lipopolysaccharide-induced dendritic cell activation and function. J. Immunol. 180, 4697–4705 (2008).
Mancino, A. et al. Divergent effects of hypoxia on dendritic cell functions. Blood 112, 3723–3734 (2008).
Yang, M. et al. HIF-dependent induction of adenosine receptor A2b skews human dendritic cells to a Th2-stimulating phenotype under hypoxia. Immunol. Cell. Biol. 88, 165–171 (2010).
Tang, C. et al. Th type 1-stimulating activity of lung macrophages inhibits Th2-mediated allergic airway inflammation by an IFN-gamma-dependent mechanism. J. Immunol. 166, 1471–1481 (2001).
Careau, E. & Bissonnette, E.Y. Adoptive transfer of alveolar macrophages abrogates bronchial hyperresponsiveness. Am. J. Respir. Cell. Mol. Biol. 31, 22–27 (2004).
Gasser, O. & Schifferli, A. Jr Activated polymorphonuclear neutrophils disseminate anti-inflammatory microparticles by ectocytosis. Blood 104, 2543–2548 (2004).
Maffia, P.C. et al. Neutrophil elastase converts human immature dendritic cells into transforming growth factor-beta1-secreting cells and reduces allostimulatory ability. Am. J. Pathol. 171, 928–937 (2007).
Eken, C., Gasser, O., Zenhaeusern, G., Oehri, I., Hess, C. & Schifferli, J.A. Polymorphonuclear neutrophil-derived ectosomes interfere with the maturation of monocyte-derived dendritic cells. J. Immunol. 180, 817–824 (2008).
Lamblin, C. et al. Bronchial neutrophilia in patients with noninfectious status asthmaticus. Am. J. Respir. Crit. Care Med. 157, 394–402 (1998).
Foley, S.C. & Hamid, Q. Images in allergy and immunology: neutrophils in asthma. J. Allergy Clin. Immunol. 119, 1282–1286 (2007).
Nabe, T. et al. Important role of neutrophils in the late asthmatic response in mice. Life Sci. 88, 1127–1135 (2011).
Denning, T.L., Wang, Y.-C., Patel, S.R., Williams, I.R. & Pulendran, B. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T cell responses. Nat. Immunol. 8, 1086–1094 (2007).
Ryan, H.E. et al. Hypoxia-inducible factor-1alpha is a positive factor in solid tumor growth. Cancer Res. 60, 4010–4015 (2000).
Clausen, B.E., Burkhardt, C., Reith, W., Renkawitz, R. & Forster, I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 8, 265–277 (1999).
Acknowledgements
We thank M Pichavant (Institut Pasteur de Lille, Lille, France) for providing endotoxin-free OVA-FITC and K Thielemans (Medical School of the Vrije Universiteit Brussel, Brussels, Belgium) for providing recombinant GM-CSF. We also thank S Ormenese and R. Stefan of the Cell Imaging and Flow Cytometry GIGA Technological Platform for FACS analyses, V. Conrath and L. Duwez from the Animal Facility for animal management, and R Fares, C François, F Olivier, J Parisi, F Perin, and I Sbai for technical and secretarial assistance. M.T. is supported by a FRIA grant (Fonds pour la Recherche dans l’Industrie et l’Agriculture) of the Wallonia-Brussels Federation of Belgium. C.J.D is supported by an FRSM grant of the FRS-FNRS (Fonds de la Recherche Scientifique Médicale, Fonds pour la Recherche Scientifique-Fonds National de la Recherche Scientifique) of the Wallonia-Brussels Federation of Belgium. The Laboratory of Cellular and Molecular Immunology is supported by grants of the FRS-FNRS (Mandat d’Impulsion Scientifique and FRSM grants) and by the Belgian Programme on Interuniversity Attraction Poles (IUAP; FEDIMMUNE) initiated by the Belgian State (Belgian Science Policy).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declared no conflict of interest
Rights and permissions
About this article
Cite this article
Toussaint, M., Fievez, L., Drion, PV. et al. Myeloid hypoxia-inducible factor 1α prevents airway allergy in mice through macrophage-mediated immunoregulation. Mucosal Immunol 6, 485–497 (2013). https://doi.org/10.1038/mi.2012.88
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mi.2012.88
This article is cited by
-
Respiratory sensitization: toxicological point of view on the available assays
Archives of Toxicology (2018)
-
Host DNA released by NETosis promotes rhinovirus-induced type-2 allergic asthma exacerbation
Nature Medicine (2017)