Abstract
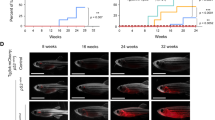
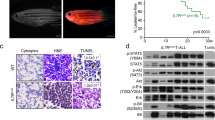
Interferon regulatory factor (IRF)-8 is a critical transcription factor involved in the pathogenesis of myeloid neoplasia. However, the underlying mechanisms in vivo are not well known. Investigation of irf8-mutant zebrafish in this study indicated that Irf8 is evolutionarily conserved as an essential neoplastic suppressor through tight control of the proliferation and longevity of myeloid cells. Surviving irf8 mutants quickly developed a myeloproliferative neoplasm (MPN)-like disease with enhanced output of the myeloid precursors, which recurred after transplantation. Multiple molecules presented notable alteration and Mertk signaling was aberrantly activated in the hematopoietic cells in irf8 mutants. Transgenic mertk overexpression in Tg(coro1a:mertk) zebrafish recapitulated the myeloid neoplasia-like syndrome in irf8 mutants. Moreover, functional interference with Mertk, via morpholino knockdown or genetic disruption, attenuated the myeloid expansion phenotype caused by Irf8 deficiency. Therefore, Mertk signaling is a critical downstream player in the Irf8-mediated regulation of the progression of myeloid neoplasia. Our study extends the understanding of the mechanisms underlying leukemogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Paun A, Pitha PM . The IRF family, revisited. Biochimie 2007; 89: 744.
Tailor P, Tamura T, Ozato K . The BXH2 mutation in IRF8 differentially impairs dendritic cell subset development in the mouse. Blood 2008; 111: 1942.
Holtschke T, Löhler J, Kanno Y, Fehr T, Giese N, Rosenbauer F et al. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell 1996; 87: 307–317.
Turcotte K, Gauthier S, Tuite A, Mullick A, Malo D, Gros P . A mutation in the Icsbp1 gene causes susceptibility to infection and a chronic myeloid leukemia-like syndrome in BXH-2 mice. J Exp Med 2005; 201: 881–890.
Schmidt M, Nagel S, Proba J, Thiede C, Ritter M, Waring JF et al. Lack of interferon consensus sequence binding protein (ICSBP) transcripts in human myeloid leukemias. Blood 1998; 91: 22–29.
Hao SX, Ren R . Expression of interferon consensus sequence binding protein (ICSBP) is downregulated in Bcr-Abl-induced murine chronic myelogenous leukemia-like disease, and forced coexpression of ICSBP inhibits Bcr-Abl-induced myeloproliferative disorder. Mol Cell Biol 2000; 20: 1149–1161.
Tamura T, Kong HJ, Tunyaplin C, Tsujimura H, Calame K, Ozato K . ICSBP/IRF-8 inhibits mitogenic activity of p210 Bcr/Abl in differentiating myeloid progenitor cells. Blood 2003; 102: 4547–4554.
Scheller M, Schoenheit J, Zimmermann K, Leser U, Rosenbauer F, Leutz A . Cross talk between Wnt/beta-catenin and Irf8 in leukemia progression and drug resistance. J Exp Med 2013; 210: 2239–2256.
Schmidt M, Bies J, Tamura T, Ozato K, Wolff L . The interferon regulatory factor ICSBP/IRF-8 in combination with PU.1 up-regulates expression of tumor suppressor p15(Ink4b) in murine myeloid cells. Blood 2004; 103: 4142–4149.
Hara T, Schwieger M, Kazama R, Okamoto S, Minehata K, Ziegler M et al. Acceleration of chronic myeloproliferation by enforced expression of Meis1 or Meis3 in Icsbp-deficient bone marrow cells. Oncogene 2008; 27: 3865–3869.
Burchert A, Cai D, Hofbauer LC, Samuelsson MK, Slater EP, Duyster J et al. Interferon consensus sequence binding protein (ICSBP; IRF-8) antagonizes BCR/ABL and down-regulates bcl-2. Blood 2004; 103: 3480–3489.
Gurevich RM, Rosten PM, Schwieger M, Stocking C, Humphries RK . Retroviral integration site analysis identifies ICSBP as a collaborating tumor suppressor gene in NUP98-TOP1 -induced leukemia. Exp Hematol 2006; 34: 1192–1201.
Cummings CT, Deryckere D, Earp HS, Graham DK . Molecular pathways: MERTK signaling in cancer. Clin Cancer Res 2013; 19: 5275.
Graham DK, Deryckere D, Davies KD, Earp HS . The TAM family: phosphatidylserine-sensing receptor tyrosine kinases gone awry in cancer. Nat Rev Cancer 2014; 14: 769–785.
Lee-Sherick AB, Eisenman KM, Sather S, McGranahan A, Armistead PM, McGary CS et al. Aberrant Mer receptor tyrosine kinase expression contributes to leukemogenesis in acute myeloid leukemia (vol 32, pg 5359, 2013). Oncogene 2016; 35: 6270–6270.
Krause S, Pfeiffer C, Strube S, Alsadeq A, Fedders H, Vokuhl C et al. Mer tyrosine kinase promotes the survival of t(1;19)-positive acute lymphoblastic leukemia (ALL) in the central nervous system (CNS). Blood 2015; 125: 820.
Minson KA, Smith CC, Deryckere D, Libbrecht C, Lee-Sherick AB, Huey MG et al. The MERTK/FLT3 inhibitor MRX-2843 overcomes resistance-conferring FLT3 mutations in acute myeloid leukemia. JCI Insight 2016; 1: e85630.
Liu J, Zhang W, Stashko MA, Deryckere D, Cummings CT, Hunter D et al. UNC1062, a new and potent Mer inhibitor. Eur J Med Chem 2013; 65: 83–93.
Langenau DM, Traver D, Ferrando AA, Kutok JL, Aster JC, Kanki JP et al. Myc-induced T cell leukemia in transgenic zebrafish. Science 2003; 299: 887–890.
Chen J, Jette C, Kanki JP, Aster JC, Look AT, Griffin JD . NOTCH1-induced T-cell leukemia in transgenic zebrafish. Leukemia 2007; 21: 462–471.
Ridges S, Heaton WL, Joshi D, Choi H, Eiring A, Batchelor L et al. Zebrafish screen identifies novel compound with selective toxicity against leukemia. Blood 2012; 119: 5621.
Sun J, Liu W, Li L, Chen J, Wu M, Zhang Y et al. Suppression of Pu.1 function results in expanded myelopoiesis in zebrafish. Leukemia 2013; 27: 1913.
Kaufman CK, White RM, Zon L . Chemical genetic screening in the zebrafish embryo. Nat Protoc 2009; 4: 1422.
Xu J, Du L, Wen Z . Myelopoiesis during zebrafish early development. J Genet Genomics 2012; 39: 435–442.
Shiau CE, Kaufman Z, Meireles AM, Talbot WS . Differential requirement for irf8 in formation of embryonic and adult macrophages in zebrafish. Plos One 2015; 10: e0117513.
Li L, Jin H, Xu J, Shi Y, Wen Z . Irf8 regulates macrophage versus neutrophil fate during zebrafish primitive myelopoiesis. Blood 2011; 117: 1359.
Li L, Yan B, Shi YQ, Zhang WQ, Wen ZL . Live imaging reveals differing roles of macrophages and neutrophils during zebrafish tail fin regeneration. J Biol Chem 2012; 287: 25353–25360.
Hall C, Flores MV, Storm T, Crosier K, Crosier P . The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev Biol 2007; 7: 42.
Traver D, Paw BH, Poss KD, Penberthy WT, Lin S, Zon LI . Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat Immunol 2003; 4: 1238–1246.
Jin SW, Beis D, Mitchell T, Chen JN, Stainier DY . Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development 2005; 132: 5199–5209.
Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B . Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol 2011; 29: 699–700.
Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X et al. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res 2013; 23: 465.
Jin H, Sood R, Xu J, Zhen F, English MA, Liu PP et al. Definitive hematopoietic stem/progenitor cells manifest distinct differentiation output in the zebrafish VDA and PBI. Development 2009; 136: 647.
Le GD, Redd MJ, Colucciguyon E, Murayama E, Kissa K, Briolat V et al. Origins and unconventional behavior of neutrophils in developing zebrafish. Blood 2008; 111: 132–141.
Link V, Shevchenko A, Heisenberg CP . Proteomics of early zebrafish embryos. BMC Dev Biol 2006; 6: 1–9.
Tamura T, Yanai H, Savitsky D, Taniguchi T . The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol 2008; 26: 535.
Jin H, Li L, Xu J, Zhen F, Zhu L, Liu PP et al. Runx1 regulates embryonic myeloid fate choice in zebrafish through a negative feedback loop inhibiting Pu.1 expression. Blood 2012; 119: 5239–5249.
Leonardi E, Girlando S, Serio G, Mauri FA, Perrone G, Scampini S et al. PCNA and Ki67 expression in breast carcinoma: correlations with clinical and biological variables. J Clin Pathol 1992; 45: 416–419.
Hendzel MJ, Wei Y, Mancini MA, Van Hooser A, Ranalli T, Brinkley BR et al. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 1997; 106: 348–360.
Orkin SH, Zon LI . Hematopoiesis: an evolving paradigm for stem cell biology. Cell 2008; 132: 631–644.
Mccubrey JA, Steelman LS, Abrams SL, Bertrand FE, Ludwig DE, Bäsecke J et al. Targeting survival cascades induced by activation of Ras/Raf/MEK/ERK, PI3K/PTEN/Akt/mTOR and Jak/STAT pathways for effective leukemia therapy. Leukemia 2008; 22: 708–722.
Ge X, Wang X . Role of Wnt canonical pathway in hematological malignancies. J Hematol Oncol 2010; 3: 1–6.
Carrà G, Torti D, Crivellaro S, Panuzzo C, Taulli R, Cilloni D et al. The BCR-ABL/NF-κB signal transduction network: a long lasting relationship in Philadelphia positive Leukemias. Oncotarget 2016; 7: 66287–66298.
Toofan P, Irvine D, Hopcroft L, Copland M, Wheadon H . The role of the bone morphogenetic proteins in leukaemic stem cell persistence. Biochem Soc Trans 2014; 42: 809–815.
Morotti A, Panuzzo C, Crivellaro S, Carrà G, Torti D, Guerrasio A et al. The role of PTEN in myeloid malignancies. Hematol Rep 2011; 7: 84–87.
Kuo YH, Jing Q, Cook GJ . Regain control of p53: targeting leukemia stem cells by isoform-specificHDAC inhibition. Exp Hematol 2016; 44: 315–321.
Le H, Zhang Y, Liu H, Ma L, Jin Y, Huang Q et al. eena promotes myeloid proliferation through stimulating ERK1/2 phosphorylation in zebrafish. J Biol Chem 2008; 283: 17652–17661.
Karin M . The regulation of AP-1 activity by mitogen-activated protein kinases. J Biol Chem 1995; 270: 16483–16486.
Camenisch TD, Koller BH, Earp HS, Matsushima GK . A novel receptor tyrosine kinase, Mer, inhibits TNF-alpha production and lipopolysaccharide-induced endotoxic shock. J Immunol 1999; 162: 3498–3503.
Murayama E, Kissa K, Zapata A, Mordelet E, Briolat V, Lin HF et al. Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity 2006; 25: 963–975.
Rhodes J, Hagen A, Hsu K, Deng M, Liu TX, Look AT et al. Interplay of Pu.1 and Gata1 determines myelo-erythroid progenitor cell fate in zebrafish. Dev Cell 2005; 8: 97.
Tamplin O, Durand E, Carr L, Childs S, Hagedorn E, Li P et al. Hematopoietic stem cell arrival triggers dynamic remodeling of the perivascular niche. Cell 2015; 160: 241.
Luttun A, Verhamme P . Keeping your vascular integrity: what can we learn from fish? Bioessays 2008; 30: 418–422.
Murray PJ, Wynn TA . Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol 2011; 11: 723.
Teittinen KJ, Grönroos T, Parikka M, Rämet M, Lohi O . The zebrafish as a tool in leukemia research. Leukemia Res 2012; 36: 1082–1088.
Gutierrez A, Grebliunaite R, Feng H, Kozakewich E, Zhu S, Guo F et al. Pten mediates Myc oncogene dependence in a conditional zebrafish model of T cell acute lymphoblastic leukemia. J Exp Med 2011; 208: 1595.
Keating A, Salzberg D, Sather S, Liang X, Nickoloff S, Anwar A et al. Lymphoblastic leukemia/lymphoma in mice overexpressing the Mer (MerTK) receptor tyrosine kinase. Oncogene 2006; 25: 6092–6100.
Roser B, Jacques P, Philippe L . ERK1 and ERK2 Map kinases: specific roles or functional redundancy? Front Cell Dev Biol 2016; 4: 53.
Klinakis A, Lobry C, Abdelwahab O, Oh P, Haeno H, Buonamici S et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature 2011; 473: 230–233.
Koenigsmann J, Carstanjen D . Loss of Irf8 does not co-operate with overexpression of BCL-2 in the induction of leukemias in vivo. Leukemia Lymphoma 2009; 50: 2078–2082.
Acknowledgements
We thank D Wang and Y Liu for technical assistances. This work was supported by the National Natural Science Foundation of China (31301198, 31271568 and 31571500); The National Key Basic Research Program of China (2015CB942802); The Fundamental Research Funds for the Central Universities (XDJK2017A015); The Outstanding Youth Science Foundation of Chongqing (cstc2011jjjq10003).
Author contributions
FZ, YS, YH and L Li designed the experiments. FZ, YS, YH, YZ and LZ performed most experiments. YL and YW provided technical assistance of flow cytometry, L Li wrote the manuscript, HL, L Luo, HH and HR discussed the results and commented on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Zhao, F., Shi, Y., Huang, Y. et al. Irf8 regulates the progression of myeloproliferative neoplasm-like syndrome via Mertk signaling in zebrafish. Leukemia 32, 149–158 (2018). https://doi.org/10.1038/leu.2017.189
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2017.189
This article is cited by
-
TANGO6 regulates cell proliferation via COPI vesicle-mediated RPB2 nuclear entry
Nature Communications (2024)
-
Zebrafish: a convenient tool for myelopoiesis research
Cell Regeneration (2023)
-
Alterations in immune cell heterogeneities in the brain of aged zebrafish using single-cell resolution
Science China Life Sciences (2023)
-
Estrogens revert neutrophil hyperplasia by inhibiting Hif1α-cMyb pathway in zebrafish myelodysplastic syndromes models
Cell Death Discovery (2022)