Abstract
Many studies have tackled the existence of a genetic barrier in the Strait of Gibraltar between Iberian and North African populations, often with controversial conclusions. Here, we address this issue using a collection of Western Mediterranean populations and two dimensionality reduction methods: principal component analysis (PCA) and spatial PCA (sPCA). Our four different data sets consisted of (i) 16 polymorphic Alu insertions in 12 populations; (ii) 35 single-nucleotide polymorphisms in 13 populations; (iii) 13 short tandem repeats in 11 populations; and (iv) all 64 markers in 9 populations. In all PCA plots, South European and North African samples were visually distinguishable along the first PC. Several smaller clusters were also identifiable, especially on the African side of our geographical setting. sPCA indicated a single global structure for each of the marker sets and no local structures. These results are more compatible with a clinal distribution of allele frequencies rather than with abrupt changes, suggesting that isolation-by-distance, rather than a barrier to gene flow, is a more likely mechanism of genetic differentiation in the Western Mediterranean. An alternative/complementary explanation is progressive introgression from North African to Southwestern European populations.
Similar content being viewed by others
Main
Human population genetics in the Western Mediterranean has a long tradition, spanning from classical studies1 to whole-genome genetic analyses.2 Many of these studies have tackled the existence of a genetic barrier in the Strait of Gibraltar between Iberian and North African populations.
With only 14.3 km of ocean separating Europe and Africa at its narrowest point, some geneticists see the Strait as a bridge for cultural and genetic diffusion, a ‘melting pot’,3 whereas others see it as a barrier to gene flow. According to the latter view, obstructed navigation between the two continents or linguistic/cultural differences may have restricted gene flow in the past.4
There are plenty of enzyme- and DNA-based studies arguing in favor of either the ‘melting pot’5, 6, 7, 8 or the ‘genetic barrier’ model.9, 10, 11, 12 These studies have mostly used genetic distances, principal component analysis (PCA) and/or spatial autocorrelation analysis.
Here we search for genetic patterns in the Western Mediterranean by use of spatial PCA (sPCA), a recently developed, spatially explicit multivariate method that reduces the multidimensionality of geo-referenced genetic data like ours to a few synthetic variables.13 Unlike ordinary PCA, which we also use here, maximization in sPCA is carried out not on genetic variance alone, but rather on the product of genetic variance and spatial autocorrelation (measured by Moran’s I).14 This way, global structures (for example, clines) are disentangled from local structures (that is, strong genetic differences between neighbors) and from random noise. Highly positive eigenvalues (for example, the product of a high genetic variance explained times a high positive autocorrelation) correspond to global patterns, whereas highly negative eigenvalues (for example, the product of a high genetic variance explained times a high negative autocorrelation) indicate local patterns.
For this study, we considered three different classes of autosomal markers: polymorphic Alu insertions (PAIs), single-nucleotide polymorphisms (SNPs) and short tandem repeats (STRs; Supplementary Table 1). The PAI data as well as part of the SNP and STR data has been reported elsewhere.8, 12, 15 All genotypic data are available in the Supplementary Excel File.
The analysis was carried out on 15 geographically well-defined Western Mediterranean populations from continental Spain, South France, Morocco, Algeria and Tunisia (Figure 1; Supplementary Table 2). Participants were healthy, unrelated individuals of either sex who had their four grandparents born in the same region. Mean sample size was ∼96 for PAIs (N=1151), ∼41 for SNPs (N=531) and ∼42 for STRs (N=467). All participants gave their informed consent and the study was performed in compliance with the guidelines of the Ethical Committee of the University of Barcelona.
Approximate geographic location of the studied population samples. AS: Oviedo, Asturias, North Spain, PA: Valles Pasiegos, Cantabria, North Spain, BA: Guipuzcoa, Basque Country, North Spain, OL: Olot, Catalonia, Northeast Spain, GD: Sierra de Gredos, Avila, Central Spain, AL: La Alpujarra, Andalusia, South Spain, TO: Toulouse, South France, DO: Doukkala, Morocco, OU: Ouarzazate, Morocco, AM: Amizmiz, High Atlas, Morocco, AN: Asni, High Atlas, Morocco, KH: Khenifra, High Atlas, Morocco, BO: Bouhria, Northeast Atlas, Morocco, MZ: M’zab, Algeria, TN: Monastir, Tunisia (see also Supplementary Table 2).
The analyses were performed on four different data sets: (i) 16 PAIs in 12 populations; (ii) 35 SNPs in 13 populations; (iii) 13 STRs in 11 populations; and (iv) all 64 markers in 9 populations (Supplementary Table 2). We carried out preliminary Mantel tests16 for correlation between genetic and geographic distances (10 000 permutations) using the ade4 v1.4-17 statistical package17 (http://pbil.univ-lyon1.fr/ADE-4/) in R v2.15.1 (http://www.r-project.org/). All correlations were modest but significant (r ranging from 0.317 to 0.564; P<0.05), indicating the presence of spatial structure in the samples. We further employed two dimensionality reduction methods on the data. First, we carried out classical PCA using ade4 to gain insight into the genetic relationships among our samples. Then, we carried out sPCA with the adegenet v1.3-5 statistical package18 (http://adegenet.r-forge.r-project.org/) in R to specifically investigate the spatial pattern of genetic variation in the studied populations.
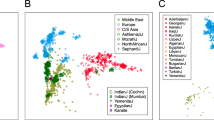
Figure 2 shows the PCA plots of the first vs second PC for the three marker sets together and separately. Linkage disequilibrium in some SNPs and STRs did not affect the results of the analysis as proven by the omission of the correlated markers (data not shown). In all plots, two clusters were visually distinguishable along the first PC, corresponding to Southwestern Europe and North Africa. Several smaller clusters were also identifiable, especially on the African side of our geographical setting. In all marker systems, the first PC explained a considerably higher percentage of genetic variance comparing with the rest of the PCs. This structure could be dating back to prehistoric times, likely to the Upper Palaeolithic, as suggested by previous age estimates for two bi-locus haplotypes (comprising of one PAI and one STR) from the same samples,8 and in agreement with the generally accepted migration pattern in the Mediterranean.19
PCA plots of the studied Western Mediterranean populations based on PAIs, SNPs and STRs together and separately. Circles correspond to South European and triangles to North African populations. Figures in brackets correspond to the percentage of variance explained by each PC. Note that plots are not in the same scale.
Despite the apparent South vs North genetic differentiation in the Western Mediterranean, PCA alone does not provide enough evidence for the existence of local structures in our sample. Such questions are better answered by methods that are based on geo-referenced data. In this light, Figure 3 shows the eigenvalues of the sPCA for the three marker sets together and separately. Because real structures tend to produce extreme positive or negative eigenvalues, sPCA indicated a single global structure for each of the marker sets and no local structures (for example, a genetic barrier between neighboring North African and South European populations). These results are more compatible with a clinal distribution of allele frequencies rather than with abrupt changes, suggesting that isolation by distance is a more likely mechanism of genetic differentiation in the Western Mediterranean. An alternative/complementary explanation is a progressive introgression from North African to Southwestern European populations, possibly reflecting the Muslim conquest of Hispania in the 8th century AC.
Bar plots of the eigenvalues of the sPCA (ordered from higher to lower) based on PAIs, SNPs and STRs together and separately. Positive eigenvalues (on the left) correspond to global structures, whereas negative eigenvalues (on the right) indicate local patterns. Actual structures should result in more extreme (positive or negative) eigenvalues. Moran’s I for PAIs: (0.617, 0.227, 0.144, 0.017, −0.082, −0.187, −0.131, −0.262, −0.400, −0.329, −0.372); SNPs: (0.569, 0.337, 0.174, 0.048, −0.038, −0.167, −0.108, −0.315, −0.275, −0.379, −0.269, −0.378); STRs: (0.427, 0.356, 0.213, −0.106, −0.106, −0.286, −0.350, −0.366, −0.272, −0.366); all markers: (0.486, 0.166, 0.073, −0.201, −0.270, −0.298, −0.342, −0.495).
Our observations contradict previous results that were arguing in favor of a major genetic barrier between the Iberian Peninsula and the Western Maghreb. Such studies were either tracing patterns of maximal genetic differentiation—typically pairwise FST values—on a specific spatial distribution provided by the geographic coordinates of the studied populations10 or they were testing for a single specific case of local structure: the one corresponding to a genetic barrier imposed by the Mediterranean Sea.12 Compared with those methods, sPCA is a more sensitive approach because (i) it takes into consideration an unbiased estimate of geographical structure (Moran’s I) and (ii) it is hypothesis free.
It is worth noting that all three data sets provided very similar results, despite the differences in the underlying mutation models. In accord with our observation, a previous study using PAIs, SNPs and STRs found high and statistically significant correlations between pairwise population genetic distances for the three marker types and for 27 worldwide populations from Africa, Asia and Europe.20
In conclusion, our analysis suggests that population structure in the Western Mediterranean is most probably the result of an isolation-by-distance mechanism between South European and North African populations. No strong local structures seem to affect the genetic landscape in the studied region. Future implementations of sPCA should include bigger data sets, as well as mtDNA and Y-chromosome data to provide information on sex-biased dispersal of allele frequencies in the Western Mediterranean.
References
Cavalli-Sforza, L. L., Menozzi, P. & Piazza, A. The history and geography of human genes, (Princeton University Press: Princeton, NJ, USA, 1994).
Botigué, L. R., Henn, B. M., Gravel, S., Maples, B. K., Gignoux, C. R., Corona, E. et al. Gene flow from North Africa contributes to differential human genetic diversity in southern Europe. Proc. Natl Acad. Sci. USA 110, 11791–11796 (2013).
González-Pérez, E., Via, M., Esteban, E., López-Alomar, A., Mazieres, S., Harich, N. et al. Alu insertions in the Iberian Peninsula and North West Africa - genetic boundaries or melting pot? Coll. Antropol. 27, 491–500 (2003).
Barbujani, G., Pilastro, A., De Domenico, S. & Renfrew, C. Genetic variation in North Africa and Eurasia: Neolithic demic difusion vs. Paleolithic colonisation. Am. J. Phys. Anthropol. 95, 137–154 (1994).
Kandil, M., Moral, P., Esteban, E., Autori, L., Mameli, G. E., Zaoui, D. et al. Red cell enzyme polymorphisms in Moroccans and southern Spaniards: new data for the genetic history of the western Mediterranean. Hum. Biol. 71, 791–802 (1999).
Plaza, S., Calafell, F., Helal, A., Bouzerna, N., Lefranc, G., Bertranpetit, J. et al. Joining the pillars of Hercules: mtDNA sequences show multidirectional gene flow in the western Mediterranean. Ann. Hum. Genet. 67, 312–328 (2003).
Bahri, R., Esteban, E., Moral, P. & Chaabani, H. New insights into the genetic history of Tunisians: data from Alu insertion and apolipoprotein E gene polymorphisms. Ann. Hum. Biol. 35, 22–33 (2008).
González-Pérez, E., Esteban, E., Via, M., Gayà-Vidal, M., Athanasiadis, G., Dugoujon, J. M. et al. Population relationships in the Mediterranean revealed by autosomal genetic data (Alu and Alu/STR compound systems). Am. J. Phys. Anthropol. 141, 430–439 (2010).
Bosch, E., Calafell, F., Pérez-Lezaun, A., Comas, D., Mateu, E. & Bertranpetit, J. Population history of North Africa: Evidence from classical genetic markers. Hum. Biol. 69, 295–311 (1997).
Comas, D., Calafell, F., Benchemsi, N., Helal, A., Lefranc, G., Stoneking, M. et al. Alu insertion polymorphisms in NW Africa and the Iberian Peninsula: evidence for a strong genetic boundary through the Gibraltar Straits. Hum. Genet. 107, 312–319 (2000).
Harich, N., Esteban, E., Chafik, A., López-Alomar, A., Vona, G. & Moral, P. Classical polymorphisms in Berbers from Moyen Atlas (Morocco): genetics, geography, and historical evidence in the Mediterranean peoples. Ann. Hum. Biol. 29, 473–487 (2002).
Athanasiadis, G., González-Pérez, E., Esteban, E., Dugoujon, J. M., Stoneking, M. & Moral, P. The Mediterranean Sea as a barrier to gene flow: evidence from variation in and around the F7 and F12 genomic regions. BMC Evol. Biol. 10, 84 (2010).
Jombart, T., Devillard, S., Dufour, A. B. & Pontier, D. Revealing cryptic spatial patterns in genetic variability by a new multivariate method. Heredity 101, 92–103 (2008).
Moran, P. The interpretation of statistical maps. J. R. Stat. Soc. Ser. B 10, 243–251 (1948).
Athanasiadis, G., Esteban, E., Gayà-Vidal, M., Dugoujon, J. M., Moschonas, N., Chaabani, H. et al. Different evolutionary histories of the Coagulation Factor VII gene in human populations? Ann. Hum. Genet. 74, 34–45 (2010).
Mantel, N. Detection of disease clustering and a generalized regression approach. Cancer Res. 27, 209–220 (1967).
Chessel, D., Dufour, A. B. & Thioulouse, J. The ade4 package- I: one-table methods. R News 4, 5–10 (2004).
Jombart, T. & Ahmed, I. adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27, 3070–3071 (2011).
Bar-Yosef, O. The upper paleolithic revolution. Annu. Rev. Antropol. 31, 363–393 (2002).
Xing, J., Watkins, W.S., Witherspoon, D.J., Zhang, Y., Guthery, S.L., Thara, R. et al. Fine-scaled human genetic structure revealed by SNP microarrays. Genome Res. 19, 815–825 (2009).
Acknowledgements
The authors would like to thank two anonymous reviewers for their useful and constructive comments on the manuscript. This study has been financially supported by the CGL2008-03955 and CGL2011-27866 projects of the Spanish Ministerio de Educación y Ciencia, as well as the 2005SGR00252 project of the Generalitat de Catalunya. The work of GA has been financed by an FPU grant from the Ministerio de Educación y Ciencia (reference: AP2005-4425). The authors would also like to thank all the donors for providing blood samples and the collaborators for collecting them: F Luna, MS Mesa and C Rodríguez (Spanish samples); N Harich and M Kandil (Moroccan samples); H Chaabani (Tunisian sample), JM Dugoujon (North African and French samples); and, two colleagues: M Gayà-Vidal for helping in the SNP genotyping and E González-Pérez for providing the PAI dataset. Also thanks to Professor M Stoneking for his advice, technical and material support during GA’s visit at the Max Planck Institute for Evolutionary Anthropology in Leipzig (Germany), where most of the STR genotyping was carried out. Finally, the authors would like to thank Professor WH Stone and ME Weale for reviewing and discussing the material presented here.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Athanasiadis, G., Moral, P. Spatial principal component analysis points at global genetic structure in the Western Mediterranean. J Hum Genet 58, 762–765 (2013). https://doi.org/10.1038/jhg.2013.94
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2013.94
Keywords
This article is cited by
-
A survey of sub-Saharan gene flow into the Mediterranean at risk loci for coronary artery disease
European Journal of Human Genetics (2017)