Abstract
An actinomycete strain S16–07T, isolated from surface-sterilized stems of rice plant (Oryza sativa L.), was characterized using a polyphasic approach. Phylogenetic analysis of 16S rRNA gene sequences indicated affiliation of the strain belonged to the genus Streptomyces. The highest levels of sequence similarity were found with Streptomyces smyrnaeus SM3501T (97.7% similarity), S. abikoensis NBRC 13860T (97.6% similarity) and S. thermocarboxydovorans NBRC 16324T (97.5% similarity). The cell wall of strain S16–07T contained LL-diaminopimelic acid. The predominant menaquinones were MK-9(H6) and MK-9(H8). Phospholipids detected were phosphatidylethanolamine, phosphatidylglycerol, diphosphatidylglycerol, hydroxy-phosphatidylethanolamine, hydroxy-phosphatidylmonomethylethanolamine and phosphatidylinositol mannosides. The major cellular fatty acids were ai-C15:0, i-C16:0 and ai-C17:0. The G+C content of strain S16–07T was 70.4 mol%. On the basis of the phylogeny of the isolate and its differences from the most closely related species, the isolate S16–07T represents a novel species for which the name S. oryzae sp. nov. is proposed. The type strain is S16–07T (=BCC 60400T=NBRC 109761T).
Similar content being viewed by others
Introduction
The genus Streptomyces was first proposed by Waksman and Henrici1 and was classified in the family Streptomycetaceae, order Actinobacteria.2 This genus currently contains more than 650 recognized species with validly published names.3 Members of the genus Streptomyces are aerobic, Gram-stain-positive, chemo-organotrophic actinomycetes and form an extensively branched substrate mycelium with rarely fragment. The aerial mycelium forms chains of three to many spores at maturity. Chemotaxonomic characteristics show lack of mycolic acids, type I cell wall containing LL-diaminopimelic acid but no characteristic sugars and contain major amounts of saturated, iso- and anteiso-fatty acids. The major menaquinone is either hexa- or octahydrogenated with nine isoprene units. The phospholipid patterns typically contain diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylinositol and phosphatidylinositol mannosides. The DNA G+C content is in the range 66–78 mol%.4 During study the diversity of endophytic actinomycetes from stems and roots of rice plant, an actinomycete designated S16–07T, was isolated and assigned to the genus Streptomyces. The aim of the present study was to determine the taxonomic status of the strain S16–07T using genotypic and phenotypic characteristics. The resultant data indicated that the organism should be classified as a novel species of Streptomyces, for which the name Streptomyces oryzae sp. nov. is proposed.
Materials and Methods
Strain S16–07T was isolated from surface-sterilized stems of rice plant (O. sativa L.) collected from Petchaburi province, Thailand. Stem samples were surface-sterilized according to the method as described by Mingma et al.5 and crushed with sterile glass rod in 1/4 strength Ringer’s solution. Crushed plant tissue suspensions were spread on starch casein agar6 supplemented with ketoconazole (100 μg ml−1), nystatin (50 μg ml−1) and nalidixic acid (25 μg ml−1) and incubated at 28 °C for 21 days. Pure colony of strain S16–07T was kept on glucose yeast extract agar (containing glucose 1.0% (w/v), yeast extract 1.0% (w/v) and agar 1.5% (w/v)). Spore and cell suspensions were stored as lyophilized ampules at 4 °C and in 20% (v/v) glycerol at −20 °C.
Strain S16–07T and the type strains of S. abikoensis NBRC 138607, S. thermocarboxydovorans NBRC 163248 and S. lilacinus ISP 52549 were studied together for biochemical, cultural and physiological characteristics. S. smyrnaeus SM3501T has recently been described by Tatar et al.10 and showed the highest similarity with the strain S16–07T. At this time of writing, S. smyrnaeus SM3501T was just published in International Journal of Systematic and Evolutionary Microbiology. Therefore, the cultural and physiological properties of S. smyrnaeus SM3501T were obtained from Tatar et al.10 for comparison purpose. Morphological observations of spores and mycelia of strain S16–07T were examined after incubation at 27 °C for 14 days by light microscope and scanning electron microscope (JEOL–JSM 5600 LV, Tokyo, Japan). Cultural characteristics were determined from the growth on ISP (International Streptomyces Project) media11 2, 3, 4 and 5, potato dextrose agar (PDA), Czapek’s agar and nutrient agar.12 The characteristics were recorded after 14 days of incubation at 27 °C. The Color Harmony Manual Charts13 were used to determine color designations. The utilization of carbohydrates as sole carbon source was investigated on ISP medium 9.11 Tolerance of sodium chloride (0, 1, 2, 3, 4, 5, 10 and 15%, (w/v)) was tested using ISP medium 2. To determine the optimal temperature and pH for growth, strain S16–07T was incubated for 14 days on ISP medium 2 at temperatures of 5–50 °C, and at pHs ranging from 3.0 to 11.0 (at intervals of 1.0 pH unit). Enzyme activity profiles were carried out using the API ZYM (bioMérieux) test kits.14 Melanin pigment was examined on ISP medium 6 and ISP medium 7.11 The production of hydrogen sulfide was detected using lead acetate strips. Hydrolysis of adenine, casein, cellulose, chitin, guanine, hypoxanthine, tyrosine, starch, xanthine and urea was examined by following the methods of Gordon and Mihm15 and Gordon et al.16
Cells for the chemotaxonomic studies were obtained after incubation of the strain in ISP medium 2 broth at 27 °C for 2 weeks in shake flasks. The cells were harvested using centrifuged and washed three times with distilled water before freeze-drying. The isomer of diaminopimelic acid was identified using the method of Becker et al.17 and Hasegawa et al.18 The sugar compositions of whole-cell wall were determined by chromatography as described by Lechevalier and Lechevalier.19 The acyl type of the cell wall was analysed by using the method of Uchida and Aida.20 Polar lipids were examined using two-dimensional TLC and identified by the method of Minnikin et al.21 The presence of mycolic acid was detected by TLC according to the method of Tomiyasu.22 Menaquinones were extracted from freeze-dried biomass using the procedure of Collins et al.23 and subsequently analyzed by LC/MS (JMS–T100LP, JEOL) with PEGASIL ODS column (2ø × 50 mm) using methanol/2-propanol (7:3). Fatty acid methyl esters were prepared and separated using a previously described by Sasser24 and identified using with the MIDI Sherlock Microbial Identification System (Microbial ID; MIDI Version 6.1). The fatty acid analysis was performed at the Faculty of Science, King Mongkut’s Institute of Technology Ladkrabang (KMITL), Thailand.
Genomic DNA was extracted as described by Kieser et al.25 and used as templates for PCR amplification and sequencing according to the procedure of Mingma et al.5 The resultant 16S rDNA sequence was aligned with closely related 16S rRNA gene sequence from the EzTaxon–e server (http://eztaxon–e.ezbiocloud.net/).26 Multiple sequence alignments were performed using the CLUSTAL W program integrated in the Molecular Evolutionary Genetics Analysis (MEGA) version 5.0.27 For phylogenetic analysis, reference strains were chosen according to the highest pairwise similarity among the top 22 BLASTN hits against the EzTaxon–e database. Phylogenetic tree was constructed by the neighbor-joining,28 maximum likelihood29 and maximum parsimony30 methods with the MEGA 5.0 software package. A phylogenetic tree and distance matrix were reconstructed by using the neighbor-joining method and generated using the models by Jukes and Cantor31. The topology of the phylogenetic tree was evaluated by the bootstrap resampling method of Felsenstein32 with 1000 replicates. Genomic DNA for hybridization was prepared according to the method described by Saito and Miura.33 DNA–DNA relatedness was measured fluorometrically using the microplate hybridization method.34 The G+C content (mol%) of the DNA was determined by HPLC according to the method of Tamaoka and Komagata.35
Results and Discussion
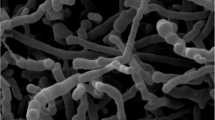
Strain S16–07T produced branched, nonfragmented substrate mycelium. The aerial mycelia harbored spore chains of hooks, open loops or primitive spiral, which belong to retinaculum apertum type of morphology that consisted of 10 or more spores per chain. Spore chain morphology of strain S16–07T was different from other closely related species as follow: S. smyrnaeus SM3501T produced spiral spore chains;10 S. abikoensis NBRC 13860T produced straight spore chains;7 S. thermocarboxydovorans NBRC 16324T formed spores in long straight to flexuous8 and S. lilacinus ISP 5254T produced verticillate spore chain.9 The spores of strain S16–07T were oval to rod shaped and 0.5 × 1.0 μm in size. Spore surface was smooth (Figure 1). The cultural characteristics of strain S16–07T on different kinds of media are presented in Table 1. Strain S16–07T showed good growth on ISP medium 2, ISP medium 5 and Czapek’s agar and moderate growth on several media including ISP medium 3, ISP medium 4, nutrient agar and PDA. The substrate mycelium of strain on most media tested was white to pale yellow with whitish aerial spore mass. Pale yellow to yellow diffusible pigment was detected when the strain was cultured on ISP medium 2, ISP medium 3, ISP medium 4 and PDA. Melanin pigment was not observed on both ISP medium 6 and ISP medium 7.
The physiological properties that differentiated strain S16–07T from S. smyrnaeus SM3501T, S. abikoensis NBRC 13860T, S. thermocarboxydovorans NBRC 16324T and S. lilacinus ISP 5254T are shown in Table 2. Strain S16–07T utilized adonitol, D(−)rhamnose, raffinose and sucrose, whereas S. abikoensis NBRC 13860T, S. thermocarboxydovorans NBRC 16324T and S. lilacinus ISP 5254T did not. In addition, S16–07T could use L(+)arabinose, D(+)cellobiose, D(+)galactose, beta-lactose and D(−)mannitol, a property which was negative in S. abikoensis NBRC 13860T and S. lilacinus ISP 5254T. S. smyrnaeus SM3501T utilized D(−)sorbitol and xylitol as the sole carbon source but strain S16–07T could not. Strain S16–07T could utilize urea and degrade casein, whereas S. smyrnaeus SM3501T and S. abikoensis NBRC 13860T could not. Strain S16–07T could tolerate NaCl at concentration up to 10%, whereas S. smyrnaeus SM3501T could tolerate up to 20%. On the other hand, S. abikoensis NBRC 13860T, S. thermocarboxydovorans NBRC 16324T and S. lilacinus ISP 5254T could tolerate only 5% NaCl. Strain S16–07T utilized fructose, glycerol, myo-inositol, maltose, trehalose and xylose as the sole carbon source but not melibiose, D(−)sorbitol and sorbose. Alkaline phosphatase, leucine aminopeptidase, acid phosphatase, phosphoamidase, α-glucosidase and glucosaminidase were detected with the API ZYM enzyme assay, but not chymotrypsin, cystine aminopeptidase, esterase (C4), esterase lipase (C8), α-fucosidase, α-galactosidase, β-galactosidase, β-glucosidase, β-glucuronidase, lipase (C14), α-mannosidase, trypsin and valine aminopeptidase. The temperature range for growth of strain S16–07T was 14–43 °C, with the optimum temperature 23–32 °C. The pH range for growth was 6.0–9.0.
An analysis of whole-cell hydrolysates showed that the strain S16–07T contained LL-diaminopimelic acid, which was characteristic for the genus Streptomyces. The whole-cell sugars were detected as galactose, glucose and ribose. The muramic acid in the peptidoglycan was N-acetylated. Polar lipids were of type II, according to the phospholipid classification of Lechevalier et al.36 and included phosphatidylethanolamine, phosphatidylglycerol, diphosphatidylglycerol, hydroxy-phosphatidylethanolamine, hydroxy-phosphatidylmonomethylethanolamine and phosphatidylinositol mannosides (Supplementary Figure S1). The major menaquinones found were MK–9(H6) (49.6%) and MK–9(H8) (41.6%), with minor amounts of MK–10(H6) (4.9%) and MK–10(H8) (3.9%). The fatty acids were ai-C15:0 (29.4%), i-C16:0 (28.3%), ai-C17:0 (17.9%), i-C15:0 (3.8%), i-C14:0 (3.8%), C16:0 (3.2%), i-H-C16:1 (3.1%), ai-C17:1 w9c (2.7%), i-C 17:0 (1.7%), i-C18:0 (0.8%), C17:0 cyclo (0.8%), ai-C13:0 (0.7%), 2OH-C17:0 (0.5%), C14:0 (0.4%), C15:1 w6c (0.4%), i-C17:1 w5c (0.4%), C17:0 (0.3%), ai-C16:0 (0.3%), i-3OH-C16:0 (0.2%) and ai-C14:0 (0.2%). Mycolic acids were not detected. The G+C content of the DNA was 70.4%.
The 16S rRNA gene-based tree, constructed using the neighbor-joining method, showed that the strain S16–07T formed a separate phyletic line from other representatives of the genus Streptomyces (Figure 2), notably from its closely related species S. smyrnaeus SM3501T (97.7%; 33/1414), S. abikoensis NBRC 13860T (97.6%; 34/1403), S. thermocarboxydovorans NBRC 16324T (97.5%; 35/1410) and S. lilacinus ISP 5254T (97.5%; 35/1404). However, it was clear that strain S16–07T represented a novel subline within the genus Streptomyces as its position in the tree was separated from these nearest phylogenetic neighbors. DNA–DNA hybridization tests were carried out between strain S16–07T and closely related strains selected on the basis of their 16S rDNA sequence similarity. DNA–DNA relatedness values between strain S16–07T and S. abikoensis NBRC 13860T (3.0%), S. thermocarboxydovorans NBRC 16324T (6.5%) and S. lilacinus ISP 5254T (13.8%) were all significantly lower than 70%, the threshold value for the delineation of genomic species.37 On the basis of 16S rRNA gene sequence data, DNA–DNA hybridization studies and biochemical properties, as well as physiological properties, it is proposed that strain S16–07T represents a novel species of genus Streptomyces, for which the name S. oryzae is proposed. The type strain is S16–07T.
Neighbor-joining phylogenetic tree, based on nearly complete 16S rRNA gene sequences, showing the relationships between strain S16–07T and strains of related species of the genus Streptomyces. Numbers at nodes are bootstrap values based on 1000 resamplings (only values >50% are indicated). Asterisks indicate that the clades are also recovered in maximum-likelihood and maximum-parsimony trees. Bar, 0.005% sequence divergence.
Description of S. oryzae sp. nov.
S. oryzae (o.ry'zae. L. gen. n. oryzae of rice, referring to the rice plant where the strain was isolated) is aerobic, Gram-stain-positive, catalase- and oxidase-positive. The substrate mycelium does not fragment. The aerial hyphae bearing smooth-surfaced spores in hooks, open loops or primitive spiral spore chain (retinaculum apertum). White aerial mycelium and white to pale yellow substrate mycelium are produced on most media. A yellow soluble pigment is produced on ISP medium 2, ISP medium 3, ISP medium 4 and PDA. Good growth occurs on ISP medium 2, ISP medium 5 and Czapek’s agar. Moderate growth is observed on ISP medium 3, ISP medium 4, nutrient agar and PDA. Melanin pigment is not produced. Growth occurs between 14 and 43 °C and at pH 6.0–9.0. Uses adonitol, L(+)arabinose, D(+)cellobiose, D(−)fructose, D(+)galactose, D(+)glucose, glycerol, myo-inositol, beta-lactose, maltose, D(−)mannitol, D(−)rhamnose, raffinose, sucrose, D(+)trehalose and xylose as sole carbon sources, but not melibiose, D(−)sorbitol, sorbose and xylitol. The organism degrades casein, hypoxanthine, starch, tyrosine, xanthine and urea, but does not degrade adenine, cellulose, chitin and guanine. Tests for nitrate reductase and H2S production are positive. LL-diaminopimelic acid is the diagnostic amino acid in the peptidoglycan and the muramic acid acyl type is acetyl. Galactose, glucose and ribose are found in whole-cell hydrolysates. The predominant menaquinones are MK-9(H6) and MK-9(H8). The major cellular fatty acids are ai-C15:0, i-C16:0 and ai-C17:0. The polar lipids include phosphatidylglycerol, diphosphatidylglycerol, hydroxy-phosphatidylethanolamine, hydroxy-phosphatidylmonomethylethanolamine, phosphatidylethanolamine and phosphatidylinositol mannosides. Mycolic acids are absent. The DNA G+C content of the type strain is 70.4 mol%.
The type strain, S16–07T (=BCC 60400T=NBRC 109761T), was isolated from stems of rice plant, O. sativa L., collected in Petchaburi province, Thailand.
Accession code
The DDBJ accession number for the 16S rRNA gene sequence of the strain S16–07T is AB894335.
References
Waksman, S. A . & Henrici, A. T . The nomenclature and classification of the Actinomycetes. J. Bacteriol. 46, 337–341 (1943).
Whitman, W. B . et al Bergey's Manual of Systematic Bacteriology: Volume 5: The Actinobacteria 2nd edn (Springer, New York, (2012).
Euzéby, J. P . List of Prokaryotic names with Standing in Nomenclature (LPSN) (2014) http://www.bacterio.cict.fr/. Accessed 29 September 2014.
Kämpfer, P . et al in Genus I. Streptomyces Waksman and Henrici 1943, 339 emend. Witt and Stackebrandt 1990, 370 emend. Wellington, Stackebrandt, Sanders, Wolstrup and Jorgensen 1992, 159. Bergey’s Manual of Systematic Bacteriology Vol. 5 (eds Goodfellow M.) 1455–1767 Springer, New York, (2012).
Mingma, R . et al. Sphaerisporangium rufum sp. nov., an endophytic actinomycete from roots of Oryza sativa L. Int. J. Syst. Evol. Microbiol. 64, 1077–1082 (2014).
Küster, E . & Williams, S. T . Selection of media for isolation of streptomycetes. Nature 202, 928–929 (1964).
Umezawa, H ., Tazaki, T . & Fukuyama, S . An antiviral substance, abikoviromycin, produced by Streptomyces species. Jpn J. Med. 4, 331–346 (1951).
Kim, S. B ., Falconer, C ., Williams, E . & Goodfellow, M . Streptomyces thermocarboxydovorans sp. nov. and Streptomyces thermocarboxydus sp. nov., two moderately thermophilic carboxydotrophic species from soil. Int. J. Syst. Bacteriol. 48, 59–68 (1998).
Nakazawa, K ., Tanabe, K ., Shibata, M ., Miyake, A . & Takewaka, T . Studies on streptomycetes. Cladomycin, a new antibiotic produced by Streptomyces lilacinus nov. sp. J. Antibiot. 9, 81 (1956).
Tatar, D ., Guven, K ., Sproer, C ., Klenk, H. P . & Sahin, N . Streptomyces iconiensis sp. nov. and Streptomyces smyrnaeus sp. nov., two halotolerant actinomycetes isolated from a salt lake and saltern. Int. J. Syst. Evol. Microbiol. 64, 3126–3133 (2014).
Shirling, E. B . & Gottlieb, D . Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340 (1966).
Waksman, S. A . The Actinomycetes. A Summary of Current Knowledge, Ronals Press, New York, (1967).
Jacobson, E ., Grauville, W. C . & Fogs, C. E . Color Harmony Manual 4th edn (Container Corporation of America, Chicago, (1958).
Humble, M. W ., King, A . & Phillips, I . API ZYM: a simple rapid system for the detection of bacterial enzymes. J. Clin. Pathol. 30, 275–277 (1977).
Gordon, R. E . & Mihm, J. M . A comparative study of some strains received as nocardiae. J. Bacteriol. 73, 15–27 (1957).
Gordon, R. E ., Barnett, D. A ., Handerhan, J. E . & Pang, C. H.-N . Nocardia coeliaca Nocardia autotrophica, and the Nocardia strain. Int. J. Syst. Bacteriol. 24, 54–63 (1974).
Becker, B ., Lechevalier, M. P . & Lechevalier, H. A . Chemical composition of cell–wall preparations from strains of various form–genera of aerobic actinomycetes. Appl. Microbiol. 13, 236–243 (1965).
Hasegawa, T ., Takizawa, M . & Tanida, S . A rapid analysis for chemical grouping of aerobic actinomycetes. J. Gen. Appl. Microbiol. 29, 319–322 (1983).
Lechevalier, H. A . & Lechevalier, M. P . in Actinomycete Taxonomy (Society for Industrial Microbiology Special Publication no. 6). The Chemotaxonomy of Actinomycetes eds Dietz A., Thayer D. W.) 277–284 VA: Society for Industrial Microbiology: Arlington, (1980).
Uchida, K . & Aida, K . Acyl type of bacterial cell wall: its simple identification by a colorimetric method. J. Gen. Appl. Microbiol. 23, 249–260 (1977).
Minnikin, D. E ., Patel, P. V ., Alshamaony, L . & Goodfellow, M . Polar lipid composition in the classification of Nocardia and related bacteria. Int. J. Syst. Bacteriol. 27, 104–117 (1977).
Tomiyasu, I . Mycolic acid composition and thermally adaptative changes in Nocardia asteroides. J. Bacteriol. 151, 828–837 (1982).
Collins, M. D ., Pirouz, T ., Goodfellow, M . & Minnikin, D. E . Distribution of menaquinones in actinomycetes and corynebacteria. J. Gen. Microbiol. 100, 221–230 (1977).
Sasser, M . Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids, MIDI Inc, Newark, DE, (1990).
Kieser, T ., Bibb, M. J ., Buttner, M. J ., Chater, K. F . & Hopwood, D. A . Practical Streptomyces Genetics, John Innes Foundation, Norwich, England, (2000).
Kim, O. S . et al. Introducing EzTaxon–e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721 (2012).
Tamura, K . et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011).
Saitou, N . & Nei, M . The neighbor–joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Felsenstein, J . PHYLIP (Phylogenetic Inference Package) Version 3.5c, Department of Genetics, University of Washington, Seattle, WA, (1993).
Fitch, W. M . Toward defining the course of evolution: minimal change for a specific tree topology. Syst. Zool. 20, 406–416 (1971).
Jukes, T. H . & Cantor, C. R in Evolution of Protein Molecules. Mammalian Protein Metabolism (ed Munro H. N.) 21–132 Academic Press, New York, (1969).
Felsenstein, J . Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791 (1985).
Saito, H . & Miura, K. I . Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim. Biophys. Acta 72, 619–629 (1963).
Ezaki, T ., Hashimoto, Y . & Yabuuchi, E . Fluorometric deoxyribonucleic acid–deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int. J. Syst. Bacteriol. 39, 224–229 (1989).
Tamaoka, J . & Komagata, K . Determination of DNA base composition by reversed–phase high–performance liquid chromatography. FEMS Microbiol. Lett. 25, 125–128 (1984).
Lechevalier, M ., Bievre, C. d . & Lechevalier, H. A . Chemotaxonomy of aerobic Actinomycetes: phospholipid composition. Biochem. Syst. Ecol. 5, 249–260 (1977).
Wayne, L. G . et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37, 463–464 (1987).
Acknowledgements
This research was supported by the Higher Education Research Promotion and National Research University Project of Thailand, Office of the Higher Education Commission; Kitasato Institute for Life Sciences, Kitasato University, Japan; Faculty of Science, Kasetsart University, Thailand.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Mingma, R., Duangmal, K., Thamchaipenet, A. et al. Streptomyces oryzae sp. nov., an endophytic actinomycete isolated from stems of rice plant. J Antibiot 68, 368–372 (2015). https://doi.org/10.1038/ja.2014.166
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2014.166
This article is cited by
-
Streptomyces spinosus sp. nov. and Streptomyces shenzhenensis subsp. oryzicola subsp. nov. endophytic actinobacteria isolated from Jasmine rice and their genome mining for potential as antibiotic producers and plant growth promoters
Antonie van Leeuwenhoek (2022)
-
Endophytic microbes: biodiversity, plant growth-promoting mechanisms and potential applications for agricultural sustainability
Antonie van Leeuwenhoek (2020)
-
Plant growth promotion by streptomycetes: ecophysiology, mechanisms and applications
Chemical and Biological Technologies in Agriculture (2016)