Abstract
Marine-derived Streptomyces sp. OPMA00072 was found to produce inhibitors of the synthesis of neutral lipids in a cell-based assay using Chinese hamster ovary (CHO) cells. A new 16-membered macrolide named bafilomycin L (BFL) (1) was isolated along with the known structurally related bafilomycin C1 (BFC1) (3) from the culture broth of the actinomycete by solvent extraction, octadecylsilyl column chromatography and HPLC. BFL inhibited cholesteryl ester (CE) synthesis in CHO cells with an IC50 value of 0.83 nM and also in mouse peritoneal macrophages with an IC50 of 6.1 nM. In addition, BFL blocked cellular acidification in HeLa cells by interfering with vacuolar H+-ATPase (V-ATPase) as well as other bafilomycins. These data strongly suggest that BFL disturbed the lysosome function to block cholesterol metabolism, leading to the inhibition of CE accumulation in mammalian cells.
Similar content being viewed by others
Introduction
Neutral lipids, cholesteryl ester (CE) and triacylglycerol (TG), are the final storage forms of cholesterol and free long-chain fatty acids in mammals.1 Cholesterol is a lipid molecule present in the membranes of all mammalian cells and is needed for their growth and viability. It is a biosynthetic precursor of bile acids, vitamin D and steroid hormones. In addition, it contributes to the development and function of the central nervous system. Excess cellular cholesterol is stored as CE. In most cell types, CE is present only at low levels, mainly as cytoplasmic lipid droplets.2 In plasma, CE is a part of the neutral lipid cargo present in the intestinal chylomicrons and in the hepatic very low-density lipoproteins (VLDL). In steroidogenic tissues such as adrenals, CE serves as the cholesterol reservoir for producing steroid hormones. However, excessive accumulation of CE in macrophages causes these to convert to foam cells, resulting in the development of atherosclerosis.3, 4, 5, 6 Furthermore, atherosclerosis can lead to serious diseases, such as myocardial and cerebral infarction.6 Therefore, inhibitors of CE accumulation are expected to be considered as therapeutic agents.
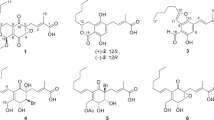
Against this background, we established a high-content cell-based assay system using Chinese hamster ovary (CHO) cells to screen microbial metabolites for inhibitors of neutral lipid accumulation. During the course of our screening program, several new microbial metabolites were discovered: verticilides7 as inhibitors of acyl-CoA:cholesterol acyltransferase and dinapinones8 and isochaetochromins9 as inhibitors of TG accumulation. From further screening studies, a new 16-membered bafilomycin congener named bafilomycin L (BFL) (1) was isolated as an inhibitor of CE synthesis from the culture broth of Streptomyces sp. OPMA00072 (Figure 1). In this study, the fermentation, isolation, structural elucidation and biological properties of BFL are described.
Results
Identification of the producing strain
In a BLAST search using BLASTn from the National Center for Biotechnology Information,10 the 16S rDNA sequence of OPMA00072 showed high levels of similarity to known Streptomyces species. Among them, the highest similarity was shown to a type strain of AY999911 (S. endus NRRL 2339; identity 1487/1508, 98.6%). Therefore the producing strain OPMA00072 was considered to belong to the genus Streptomyces.
Fermentation
Strain OPMA00072 was isolated from litter collected at the mouth of Hijya River in Okinawa. The mycelia of the strain were used for inoculation in a 500-ml Erlenmeyer flask containing 100 ml of seed medium (1.0% glucose, 0.40% polypeptone, 0.40% yeast extract, 0.050% MgSO4·7H2O and 0.10% K2HPO4). The flask was shaken on a rotary shaker at 27 °C for 7 days. The seed culture (1.0 ml) was used for inoculation in a 500-ml Erlenmeyer flask containing 100 ml of production medium (2.0% glucose, 2.0% powder of kelp tea (Gyokuroen Co., Ltd, Tokyo, Japan), and 0.90% Daigo’s artificial seawater (Nihon Pharmaceutical Co., Ltd, Tokyo, Japan), pH 7.6). The fermentation was carried out under shaking conditions at 27 °C for 9 days.
Isolation
The culture broth (5 l) was centrifuged (8000 r.p.m., 10 min) and the mycelia were collected. The cell body was treated with acetone (500 ml). After the acetone extracts were filtrated and concentrated, the resulting aqueous solution was extracted with EtOAc (1 l). The organic layer was dried over Na2SO4 and concentrated under reduced pressure to give a dark green material (845 mg). This material was dissolved in 20% MeOH and applied to an octadecylsilyl column (50 g). Active principles were eluted stepwise with 20, 40, 60, 80% MeOH aqueous and 100% MeOH (300 ml each). The 100% MeOH fraction was concentrated under reduced pressure to give an yellow material (120 mg). This material was finally purified by preparative HPLC under the following conditions: column, PEGASIL octadecylsilyl, 20 × 250 mm (Senshu Scientific Co., Tokyo, Japan); mobile phase, 80% CH3CN-0.05% trifluoroacetic acid (TFA) for 30 min, followed by a 5.0-min gradient from 80% CH3CN-0.050% TFA to 100% CH3CN-0.050% TFA; detection, UV at 247 nm; and flow rate, 8 ml min−1. Under these conditions, bafilomycin C1 (BFC1)11 and BFL were eluted as peaks with retention times of 22 and 31 min, respectively. The fractions were concentrated under reduced pressure to give pure BFC1 (2.2 mg) and BFL (16.4 mg).
Structural elucidation of BFL
The physico-chemical properties of BFL are summarized in Table 1. The molecular formula of BFL was determined to be C35H52O5 on the basis of HR-FAB-MS measurement. BFL had UV absorption at 245 and 280 nm, which was similar to that of BFC1 (247 and 287 nm). On the IR spectrum, BFL showed absorption maxima at 3427 and 1700 cm−1, suggesting the presence of hydroxy and carbonyl moieties, respectively. The 13C NMR spectrum (in DMSO-d6) showed 35 resolved signals, which were classified into 11 sp3 methyl carbons, including one methoxy carbon, one sp3 methylene carbon and 8 sp3 methine carbons, 8 sp2 methine carbons and 7 quaternary carbons, including one carbonyl carbon by analysis of DEPT. The connectivity of proton and carbon atoms was established by the HMQC spectrum, as shown in Table 2. Analysis of the 1H-1H COSY spectrum gave the four partial structures C-5 to C-9, C-29 and C-30, C-11 to C-17 and C-32, C-20 to C-21 and C-23 to C-26 (Figure 2a). The 13C-1H long-range couplings of 2J and 3J observed in the 13C-1H HMBC experiments (Figure 2a) gave the following information. (1) The cross peaks from 27-H3 (δ 1.94) to C-1 (δ 168.1), C-2 (δ 120.6) and C-3 (δ 145.9), from 28-H3 (δ 1.87) to C-3, C-4 (δ 131.7) and C-5 (δ 145.7), from 29-H3 (δ 0.94) to C-5, C-6 (δ 38.4) and C-7 (δ 78.4), from 30-H3 (δ 0.91) to C-7, C-8 (δ 37.4) and C-9 (δ 41.6), from 31-H3 (δ 1.56) to C-9, C-10 (δ 140.5) and C-11 (δ 124.1), from 14-OCH3 (δ 3.10) to C-14 (δ 82.0), from 15-H (δ 4.75) to C-1 and from 32-H3 (δ 0.88) to C-15 (δ 77.2), C-16 (δ 34.4) and C-17 (δ 128.1) supported the partial structure I. (2) The cross peaks from 33-H3 (δ 1.73) to C-18 (δ 127.6) and C-19 (δ 150.0), from 20-H (δ 5.29) to C-19, C-21 (δ 118.5) and C-22 (δ 128.5), from 34-H3 (δ 1.73) to C-21, C-22 and C-23 (δ 82.8) and from 23-H (δ 4.31) to C-19, C-21, C-24 (δ 29.9), C-25 (δ 18.7), C-26 (δ 16.8) and C-34 (δ 20.0) supported the partial structure II. (3) The cross peaks from 17-H (δ 5.92) to C-19 and from 33-H3 to C-17 indicated that the partial structures I and II are joined, as shown in Figure 2a. Taking into consideration the molecular formula, the IR data and the chemical shift of C-7 (δ 78.4), one hydroxyl moiety should be bound to C-7. Thus the planar structure of BFL was elucidated, as shown in Figure 2a.
Stereochemistry of BFL
The relative stereochemistry of BFL was determined by comparison of coupling constants and ROESY experiments with known bafilomycins.12 These data were very similar with those of BFL. The coupling constants of 11-H (11.4 Hz), 12-H (11.4 and 15.1 Hz) and 13-H (5.3 and 15.1 Hz) are supported 10E and 12E. In addition, observation of NOEs from 3-H to 5-H, from 6-H to 7-H, 8-H and 28-H3, from 7-H to 29-H3, from 8-H to 11-H, from 9-H to 30-H3, from 14-H to 14-OCH3 and 15-H and from 15-H to 32-H3 (Figure 2b) indicated that they could be assigned as 2E, 4E, 6R*, 7S*, 8S*, 14S*, 15R* and 16S*, showing the same stereochemistry as those of bafilomycins reported previously.13 However, the stereochemistry of C-23 was not elucidated by the ROESY experiments. For elucidation of the absolute stereochemistry, we need more investigations.
Biological properties
Inhibition of CE synthesis in CHO cells by bafilomycins
As shown in Table 3, BFL inhibited [14C]CE synthesis in CHO cells but had no effect on [14C]TG synthesis, even at 17.5 nM. Thus BFL was found to show selective CE inhibition with a selectivity index (SI; IC50 of TG/IC50 of CE) of >21. Under the same conditions, bafilomycin A1 (BFA1) and BFC1 also showed selective inhibition of CE synthesis with SI of 11 and >34, respectively. As a result, these bafilomycins showed similar effects on lipid metabolism in CHO cells.
Inhibition of CE synthesis in mouse peritoneal macrophages by BFL
To confirm the inhibitory activity of CE synthesis by BFL, the effect of BFL on lipid droplet formation in mouse peritoneal macrophages was investigated according to our established method.14 In this macrophage assay, about 40% of exogenously added [14C]oleic acid was incorporated into [14C]CE and [14C]TG, the main constituents of lipid droplets in the cytosol. The lipid droplets were also observed microscopically by oil red O staining.
As summarized in Table 3, BFL showed selective inhibition of CE synthesis (IC50: 6.1 nM) with an SI of>28 in this assay. BFA1 and BFC1 also exhibited similar inhibitory characteristics (SI: >43 and >35, respectively). BFL was found to dose-dependently inhibit lipid droplet accumulation (Figure 3), with marked reductions of the number and size of the lipid droplets being observed (17∼170 nM). No cytotoxic effect was observed even at 170 nM.
Inhibition of V-ATPases by BFL
It is well known that bafilomycins are potent inhibitors of vascular-type H+-ATPase (V-ATPase).15, 16, 17 As the low pH of intracellular acidic organelles like lysosomes is maintained by V-ATPase, the effect of BFL on organelle acidification was investigated. As shown in Figure 4a, acidic organelles in HeLa cells without bafilomycins (control) were stained with acridine orange. However, in the presence of BFL or BFA1, the orange fluorescence was dose-dependently faint, indicating that both compounds inhibited V-ATPase in HeLa cells.
Next, the direct effect of BFL on V-ATPase activity was tested. ATPase activity in membrane fractions prepared from Saccharomyces cerevisiae strain BY4741 was determined by the established methods using malachite green.18 As shown in Figure 4b, both BFL and BFA1 inhibited yeast V-ATPase activity with similar IC50 values of 5.3 and 2.5 nM, respectively.
Binding site of BFL on V-ATPase
Bowman et al.19 demonstrated that BFA1 binds to the V0 domain of V-ATPase in S. cerevisiae and that a T32I mutation of the c subunit (encoded by VMA3) of the V0 domain confers the strongest resistance to BFA1. To determine whether the binding site of BFL is the same as that of BFA1, the effect of BFL on the growth of vma3-T32I mutant S. cerevisiae was carried out according to an established method.18 The growth of the wild-type S. cerevisiae was inhibited by BFA1 and BFL (Figure 5), but the vma3-T32I mutant was resistant to BFL as well as BFA1. These results indicate that BFL binds to the BFA1-binding site in the c subunit.
Antimicrobial activity
No antimicrobial activity of BFL (10 μg per disk) against Bacillus subtilis, Staphylococcus aureus, Micrococcus luteus, Escherichia coli, Pseudomonas aeruginosa and Candida albicans was observed.
Discussion
In this study, we discovered BFL, a new member of the bafilomycin family, from a marine-derived Streptomyces strain. The bafilomycin family has a common 16-membered lactone ring with two conjugated diene moieties at the same positions and the same side chains at C-6, C-7, C-8 and C-14 (Figure 1). This family includes 14 bafilomycins,11, 20, 21, 22 hygrolidin,23 setamycin,24, 25 hygrobafilomycin,12 JBIR-10026 and makinolides,27, 28 all of which are produced by Streptomyces strains. As elucidated in this study, BFL has two structural characteristics: (1) a methyl residue at C-2 like makinolides, JBIR-100, bafilomycin K and J, but on an O-methyl residue at C-2 in other members, and (2) a 2H-pyran at C-18, but a tetrahydropyran or a dihydropyran at C-18 in other members.
Bafilomycins have a variety of biological activities, such as antibacterial, antifungal,11 antineoplastic,29 antiosteoporotic30 and for neuronal protection.31 As for the mechanism of action, they are well known as inhibitors of V-ATPases,15 and representative BFA1 has been utilized as a useful biochemical tool. Furuchi et al.32 reported that BFA1 induced cholesterol accumulation in the lysosomes in mouse peritoneal macrophages by blockage of V-ATPases. In their experiment, phosphatidylserine-containing liposomes with cholesterol were used as a substrate for lipid droplet formation, which were internalized via receptor-mediated endocytosis and were hydrolyzed in the lysosomes. Therefore, it makes sense that BFA1 inhibited the lysosomal function to accumulate cholesterol in the lysosomes, leading to reduction of CE synthesis and lipid droplet synthesis in the cytosol. In other studies, it was reported that bafilomycin B1 and concanamycin B, which was 18-membered macrolide isolated from Streptomyces diastatochromogenes,33 also inhibited ATP-dependent acidification of lysosomes and endosomes, resulted in the reduction of oxidized LDL-induced synthesis of CE and thereby caused a reduced accumulation of lipid droplets in macrophage J774.34, 35 The present study demonstrated that BFL showed the same action on mouse macrophages as BFA1. That is, BFL inhibited V-ATPase in HeLa cells (Figure 4) by binding to the same active site of the V0 domain as that of BFA1 (Figure 5), leading to the inhibition of CE synthesis (Table 3) and the reduction of lipid droplet formation in mouse macrophages (Figure 3). In the CHO cell assay, these bafilomycins also inhibited [14C]CE synthesis from [14C]oleic acid (Table 3). Therefore, it is plausible that cholesterol is supplied to ER, where CE synthesis occurs from free cholesterol and [14C]oleic acid or [14C]oleoyl-CoA, via V-ATPase-containing organelles, including lysosomes.
Recently, Carr et al.21 reported an intriguing action of bafilomycins on autophagy. They suggested that bafilomycin I (BFI), which has the conjugated Δ17,18, Δ19,20 diene moiety in the side chain part, stimulated autophagy, while other bafilomycins blocked autophagosomal degradation. As BFL contained the conjugated diene in the side chain like BFI, it will be worth testing whether BFL stimulates autophagy.
Materials and methods
General
Various NMR spectra were obtained using the NMR System 400 MHz and an INOVA 600 MHz spectrometer (Agilent Technologies, Santa Clara, CA, USA). FAB-MS spectra were recorded on a mass spectrometer (JMS-700 Mstation; JEOL, Tokyo, Japan). Optical rotations were measured with a digital polarimeter (DIP-1000; JASCO, Tokyo, Japan). UV spectra were recorded on a spectrophotometer (8453 UV-visible spectrophotometer; Agilent Technologies). IR spectra were recorded on a Fourier transform IR spectrometer (FT-710; Horiba Ltd, Kyoto, Japan).
Materials
[1-14C]Oleic acid (1.85 GBq mmol−1) was purchased from PerkinElmer (Waltham, MA, USA). Fetal bovine serum was purchased from Biowest (Nuaille, France). Dulbecco’s modified Eagle’s medium and Hank’s buffered salt solution were purchased from Nissui Pharmaceutical Co. (Tokyo, Japan). GIT medium was from Nippon Seiyaku Co. (Tokyo, Japan). Penicillin (10 000 units ml−1), streptomycin (10 000 mg ml−1) and glutamine (200 mM) solution were from Invitrogen (Carlsbad, CA, USA). Phosphatidylcholine, phosphatidylserine, dicetylphosphate, cholesterol, Ham’s F-12 medium, malachite green oxalate and BFA1 were purchased from Sigma-Aldrich (St Louis, MO, USA). Perchloric acid and Triton X-100 were purchased from Wako (Osaka, Japan).
Identification of Streptomyces sp. OPMA00072
For DNA extraction, the strain was cultured on yeast malt agar medium (IPS medium No. 2) at 27 °C under dark conditions for 2 weeks. An agar disc containing a mycelium was broken in TPS buffer (100 mM Tris–HCl, 10 mM EDTA, 1 M KCl; pH 8.0) and centrifuged for 5 min at 16 000 g. Total DNA was extracted by isopropanol precipitation.36 From the extracted DNA, a partial sequence of 16S rDNA was amplified by PCR. The reactions were performed as a template of TaKaRa Ex Taq (TaKaRa Bio Inc., Shiga, Japan). The amplified product was purified using a Multi Screen HTS PCR96 Filter Plate (Merck Millipore, Darmstadt, Germany) and sequenced. A partial sequence of 16S rDNA of the strain is available in the DDBJ/EMBL/GenBank databases under the accession number AB896819. Simplified identification of the strain was conducted by similarity search of the sequence using the BLASTn program on the DNA Data Bank of Japan. The sequence analyzed in this study was compared with the sequences stored in the DDBJ database.
Cell culture
CHO cells (a generous gift from Dr Kentaro Hanada, National Institute of Infectious Diseases, Tokyo, Japan) were maintained at 37 °C in 5.0% CO2 in medium A containing Ham’s F-12 medium supplemented with 10% heat-inactivated fetal bovine serum using a method described previously.37
Assay for neutral lipid synthesis in CHO cells
Assays for CE, TG and phospholipid (PL) synthesis using CHO cells were carried out by our established method.38 In brief, CHO cells (1.25 × 105 cells) were cultured in each well of a 48-well plastic microplate (Corning Co., Corning, NY, USA) in 250 μl of medium A and allowed to recover overnight at 37 °C in 5.0% CO2. The assays were carried out under at least 80% confluent cell conditions. Following overnight recovery, a test sample (in 2.5 μl of CH3CN) and [14C]oleic acid (1 nmol, 1.85 kBq in 5.0 μl of 10% EtOH/phosphate-buffered saline (PBS)) were added to each well of the culture. After 6 h of incubation at 37 °C in 5.0% CO2, cells in each well were washed twice with PBS and lysed by adding 0.25 ml of 10 mM Tris-HCl (pH 7.5) containing 0.10% (w/v) sodium dodecyl sulfate, and the cellular lipids were extracted by the method of Bligh and Dyer.39 After concentrating the organic solvent, the total lipids were separated on a TLC plate (silica gel F254, 0.5 mm thick, Merck), which was analyzed with a bioimaging analyzer (FLA-7000; Fujifilm, Tokyo, Japan) to measure the amount of [14C]CE and [14C]TG. The IC50 value was defined as a drug concentration causing 50% inhibition of each lipid synthesis.
Assay for neutral lipid synthesis in mouse peritoneal macrophages
The assay for the synthesis of neutral lipid from [14C]oleic acid was carried out according to a previously described method.14 Briefly, mouse peritoneal macrophages (5.0 × 105 cells/0.25 ml of medium B (containing Dulbecco’s modified Eagle’s medium supplemented with 8.0% (v/v) lipoprotein-deficient serum, penicillin (100 units ml−1) and streptomycin (100 mg ml−1)) were cultured in each well of a 48-well plastic microplate with a test compound (in 2.5 μl of CH3CN) and liposomes (10.0 μl, 1.0 μmol phosphatidylcholine, 1.0 μmol phosphatidylserine, 0.20 μmol dicetylphosphate and 1.5 μmol cholesterol, suspended in 1.0 ml of 0.30 M glucose) together with [14C]oleic acid (5.0 μl (1.85 kBq) in 10% ethanol/PBS solution). Following 14 h of incubation, cellular lipids were extracted to measure the radioactivity of [14C]CE and [14C]TG according to the same method as described above.
Assay for lipid droplet accumulation in mouse macrophages
The assay for lipid droplet accumulation in mouse peritoneal macrophages was carried out according to a previously described method.14 In brief, primary mouse peritoneal macrophages (5.0 × 105 cells in GIT medium) in each well of an eight-well glass chamber slide (Nalge Nunc International K.K., Rochester, NY, USA) were incubated in a humidified CO2 (5.0% v/v) atmosphere at 37 °C for 2 h. The medium was then replaced with 0.25 ml of medium B. After another 2 h of preincubation, a sample (in 2.5 μl of CH3CN) and liposomes (10.0 μl) were added to each well. After 14 h of incubation, the cells were washed with PBS and then fixed by soaking in 10% formalin. Nuclei and intracellular neutral lipid droplets were then stained with hematoxylin and oil red O, respectively. The lipid droplet formation and morphological changes in macrophages were examined by confocal-laser scanning microscopy (LSM-510 META, Carl Zeiss, Oberkochen, Germany).
Staining of intracellular acidic organelles
HeLa cells were stained with acridine orange as described previously18 with slight modification. HeLa cells were seeded onto coverslips and incubated for 2 h with BFA1 or BFL at the indicated concentrations. They were incubated for 15 min with 5 μM acridine orange, and then the coverslips were observed with a fluorescence microscope (LAS AF 6000, Leica, Wetzlar, Germany).
Vacuole isolation and measurement of V-ATPase activities
Vacuole isolation was performed as described previously with slight modification.40 In brief, S. cerevisiae BY 4741 strain was cultured on YPD medium for 9 h at room temperature. Growing cells were harvested by centrifugation at 5000 r.p.m. for 5 min at 4 °C, washed with sterile water and suspended in buffer A (1 M sorbitol, 10 mM Tris–HCl, pH 7.0) containing 1 mM dithiothreitol for 5 min at room temperature. To digest the cell wall, cells were incubated in buffer A containing 40 mg of zymolyase at 37 °C for 1 h. The mixture was washed with buffer A, resuspended in buffer A containing 12% Ficoll and homogenized with 10 strokes on ice. For the isolation of vacuoles, supernatant was transferred to ultracentrifuge tubes, and buffer B (8% Ficoll, 10 mM Tris–HCl, pH 7.0) was layered on top. The tubes were centrifuged at 18 000 r.p.m. for 60 min at 0 °C. The white material on top of the tubes was collected in new tubes, which were filled with buffer B and centrifuged at 18 000 r.p.m. for 60 min at 0 °C. After recentrifugation, the white material on top of the tubes was collected in a homogenizer and gently homogenized with five strokes. The obtained samples were used for the subsequent experiment.
V-ATPase activity assay was performed by the malachite green method as described previously.18 In brief, the reaction mixture (150 μl) contained 5 mM Na2ATP, 5 mM MgCl2, 10 mM NH4Cl, 5 mM NaN3, 0.1 mM Na3VO4, 25 mM Mes-Tris (pH 6.9) and vacuolar membrane vesicles (5 μg of protein). The mixture (without ATP) was incubated with or without a drug at the indicated concentrations for 10 min on ice, and then the reaction was started by adding ATP. Activity assays were run for 20 min at 37 °C. The reaction was stopped by adding 0.6 M perchloric acid (150 μl), and then the mixture was put on ice. The mixture was incubated with malachite green buffer containing 0.03% malachite green oxalate, 0.2% NaMoO4, 0.05% Triton X-100 and 0.7 M HCl for 40 min at room temperature. In this condition, malachite green forms a green complex with inorganic phosphate from ATP degradation. The absorbance at 650 nm was measured with a spectrometer (DU800; Beckman Coulter Inc., Brea, CA, USA).
Yeast manipulation
The effects of BFL on the growth of cells containing the wild-type VMA3 gene (SKY080; MATa his3 ura3 leu2 trp1 ade2 can1 pdr1::hisG pdr3::hisG yrs1::HIS3 yrr1::TRP1 vma3::natNT2, lue2-pRS305-VMA3) or vma3-T32I gene (ATU430; MATa his3 ura3 leu2 trp1 ade2 can1 pdr1::hisG pdr3::hisG yrs1::HIS3 yrr1::TRP1 vma3::natNT2, leu2-pRS305-vma3-T32I) were examined as follows.18 VMA3 and vma3-T32I cells were grown on YPDA medium (1.0% yeast extract, 2.0% polypeptone, 2.0% glucose and 0.020% adenine). For serial dilution assay, the cell number was estimated using the absorbance at 600 nm, and diluted to 1 × 107 cells ml−1 in water. Serial fivefold dilutions of these cells in water (3.0 μl aliquots) were pipetted onto YPDA plates (pH 7.0) containing BFA1 or BFL. These plates were incubated at 30 °C for 3 days and were photographed.
Antimicrobial activity
Antimicrobial activity of a sample against six species of microorganism was measured by the agar diffusion method using paper disks. Media for microorganisms were as follows: nutrient agar (Sanko Junyaku Co., Ltd, Tokyo, Japan) for B. subtilis PCI219, S. aureus FDA209P, M. luteus KB212, E. coli JM109 and P. aeruginosa IFO12689, and a medium composed of 1.0% glucose, 0.50% yeast extract and 0.80% agar for C. albicans ATCC90029. A paper disk (i.d. 6 mm; Toyo Roshi Kaisha, Tokyo, Japan) containing a sample (10 μg) was placed on the agar plate. Bacteria were incubated at 37 °C for 24 h. C. albicans was incubated at 27 °C for 48 h. Antimicrobial activity is expressed as the diameter (mm) of the inhibitory zone.
Accession codes
References
Athenstaedt, K. & Daum, G. The life cycle of neutral lipids: synthesis, storage and degradation. Cell Mol. Life Sci. 63, 1355–1369 (2006).
Johnson, W. J., Phillips, M. C. & Rothblat, G. H. Lipoproteins and cellular cholesterol homeostasis. Subcell. Biochem. 28, 235–276 (1997).
Goldstein, J. L., Ho, Y. K., Basu, S. K. & Brown, M. S. Binding site on macrophages that mediates uptake and degradation of acetylated low density lipoprotein, producing massive cholesterol deposition. Proc. Natl. Acad. Sci. USA 76, 333–337 (1979).
Brown, M. S., Goldstein, J. L., Krieger, M., Ho, Y. K. & Anderson, R. G. Reversible accumulation of cholesteryl esters in macrophages incubated with acetylated lipoproteins. J. Cell Biol. 82, 597–613 (1979).
Schaffner, T. et al. Arterial foam cells with distinctive immunomorphologic and histochemical features of macrophages. Am. J. Pathol. 100, 57–73 (1980).
Moore, K. J. & Tabas, I. Machophages in the pathogenesis of atherosclerosis. Cell 145, 341–355 (2011).
Ohshiro, T. et al. New verticilides, inhibitors of acyl-CoA:cholesterol acyltransferase, produced by Verticillium sp. FKI-2679. J. Antibiot. 65, 255–262 (2012).
Ohte, S. et al. Dinapinones, novel inhibitor of triacylglycerol synthesis in mammalian cells, produced by Penicillium pinophilum FKI-3864. J. Antibiot. 64, 489–494 (2011).
Ugaki, N. et al. New isochaetochromin, an inhibitor of triacylglycerol synthesis in mammalian cells, produced by Penicillium sp. FKI-4942: I. Taxonomy, fermentation, isolation and biological properties. J. Antibiot. 65, 15–19 (2012).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Warner, G. et al. Metabolic products of macroorganisms. 224. Bafilomycins, a new group of macrolide antibiotics. Production, isolation, chemical structure and biological activity. J. Antibiot. 37, 110–117 (1984).
Tchize Ndejouong Ble., S. et al. Hygrobafilomycin, a cytotoxic and antifungal macrolide bearing a unique monoalkylmaleic anhydride moiety, from Streptomyces varsoviensis. J. Antibiot. 63, 359–363 (2010).
O’Shea, M. G., Rickards, R. W., Rothschild, J. M. & Lacey, E. Absolute configurations of macrolide antibiotics of the bafilomycin and leucanicidin groups. J. Antibiot. 50, 1073–1077 (1997).
Namatame, I., Tomoda, H., Arai, H., Inoue, K. & Ōmura, S. Complete inhibition of mouse macrophage-derived foam cell formation by triacsin C. J. Biochem. 125, 319–327 (1999).
Bowman, E. J., Siebers, A. & Altendorf, K. Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc. Natl. Acad. Sci. USA 85, 7972–7976 (1988).
Drose, S. et al. Inhibitory effect of modified bafilomycins and concanamycins on P- and V-type adenosinetriphosphatases. Biochemistry 32, 3902–3906 (1993).
Wang, Y. R., Inoue, T. & Forgac, M. Subunit a of the yeast V-ATPase participates in binding of bafilomycin. J. Biol. Chem. 280, 40481–40488 (2005).
Kazami, S. et al. Iejimalides show anti-osteoclast activity via V-ATPase inhibition. Biosci. Biotechnol. Biochem. 70, 1364–1370 (2006).
Bowman, E. J., Graham, L. A., Stevens, T. H. & Bowman, B. J. The bafilomycin/concanamycin binding site in subunit c of the V-ATPases from Neurospora crassa and Saccharomyces cerevisiae. J. Biol. Chem. 279, 33131–33138 (2004).
Kretschmer, A., Drogerloh, M., Deeg, M. & Hagenmaier, H. The structures of novel insecticidal macrolides bafilomycins D and E, and oxohygrolidin. Agric. Biol. Chem. 49, 2509–2511 (1985).
Carr, G. et al. Bafilomycins produced in culture by Streptomyces spp. isolated from marine habitats are potent inhibitors autophagy. J. Nat. Prod. 73, 422–427 (2010).
Zhang, D. J. et al. Bafilomycin K, a new antifungal macrolide from Streptomyces flavotricini Y12-26. J. Antibiot. 64, 391–393 (2011).
Seto, H., Tajima, I., Akao, H., Furihata, K. & Otake, N. The isolation and structures of hygrolidin amide and defumarylhygrolidin. J. Antibiot. 37, 610–613 (1984).
Omura, S., Otoguro, K., Nishikiori, T., Oiwa, R. & Iwai, Y. Setamycin, a new antibiotic. J. Antibiot. 34, 1253–1256 (1981).
Otoguro, K., Nakagawa, A. & Omura, S. Setamycin, a 16-membered macrolide antibiotic. Identification and nematocidal activity. J. Antibiot. 41, 250–252 (1988).
Ueda, J. et al. A new 16-membered tetraene macrolide JBIR-100 from a newly identified Streptomyces species. J. Antibiot. 63, 627–629 (2010).
Kodani, S., Murao, A., Hidaki, M., Sato, K. & Ogawa, N. Isolation and structural determination of a new macrolide, makinolide, from the newly isolated Streptomyces sp. MK-30. J. Antibiot. 65, 331–334 (2012).
Hidaki, M., Yoshida, M., Ogawa, N. & Kodani, S. Isolation and structural determination of makinolide B from Streptomyces sp. MK-19. Biosci. Biotechnol. Biochem. 77, 1964–1966 (2013).
Beutler, J. A. & McKee, T. C. Novel marine and microbial natural product inhibitors of vascuolar ATPase. Curr. Med. Chem. 10, 787–796 (2003).
Xu, J., Cheng, T., Feng, H. T., Pavlos, N. J. & Zheng, M. H. Structure and function of V-ATPases in osteoclasts: potential therapeutic targets for the treatment of osteolysis. Histol. Histopathol. 22, 443–454 (2007).
Pivtoraiko, V. N. et al. Low-dose bafilomycin attenuates neuronal cell death associated with autophagy-lysosome pathway dysfunction. J. Neurochem. 114, 1193–1204 (2010).
Furuchi, T., Aikawa, K., Arai, H. & Inoue, K. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, blocks lysosomal cholesterol trafficking in macrophages. J. Biol. Chem. 268, 27345–27348 (1993).
Kinashi, H., Someno, K. & Sakaguchi, K. Isolation and characterization of concanamycins A, B, and C. J. Antibiot. 37, 1333–1343 (1984).
Naganuma, S., Kuzuya, N., Sakai, K., Hasumi, K. & Endo, A. Inhibition of the accumulation of lipid droplets in macrophage J774 by bafilomycin B1 and destruxin E. Biochim. Biophys. Acta 1126, 41–48 (1992).
Woo, J. T., Shinohara, C., Sakai, K., Hasumi, K. & Endo, A. Inhibition of the acidification of endosomes and lysosomes by the antibiotic concanamycin B in macrophage J774. Eur. J. Biochem. 207, 383–389 (1992).
Eshima, A., Wada, T., Tsubine, T. & Ogata, T. An efficient rice variety discrimination system using simple sequence repeat markers. Bull. Fukuoka Agric. Res. Cent. 26, 19–23 (2007).
Lada, A. et al. Identification of ACAT1- and ACAT2-specific inhibitors using a novel, cell-based fluorescence assay: individual ACAT uniqueness. J. Lipid Res. 45, 378–386 (2004).
Ohshiro, T., Rudel, L. L., Ōmura, S. & Tomoda, H. Selectivity of microbial acyl-CoA: cholesterol acyltransferase inhibitors toward isozymes. J. Antibiot. 60, 43–51 (2007).
Bligh, E. G. & Dyer, W. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 (1959).
Uchida, E., Ohsumi, Y. & Anraku, Y. Purification and properties of H+-translocationg, Mg2+-adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae. J. Biol. Chem. 260, 1090–1095 (1985).
Acknowledgements
We thank Ms Noriko Sato and Dr Kenichiro Nagai (School of Pharmaceutical Sciences, Kitasato University) for measurements of NMR spectra and MS data. This work was supported by a grant-in-aid for Scientific Research (A) 26253009 from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to HT).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kobayashi, K., Fukuda, T., Usui, T. et al. Bafilomycin L, a new inhibitor of cholesteryl ester synthesis in mammalian cells, produced by marine-derived Streptomyces sp. OPMA00072. J Antibiot 68, 126–132 (2015). https://doi.org/10.1038/ja.2014.100
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2014.100
This article is cited by
-
Characterization of a gamma-butyrolactone synthetase gene homologue (stcA) involved in bafilomycin production and aerial mycelium formation in Streptomyces sp. SBI034
Applied Microbiology and Biotechnology (2016)
-
Graphiumins, new thiodiketopiperazines from the marine-derived fungus Graphium sp. OPMF00224
The Journal of Antibiotics (2015)