Abstract
Platensimycin and platencin are novel antibiotics produced by Streptomyces platensis. They are potent and non-toxic natural products active against Gram-positive pathogens, including antibiotic-resistant strains and Mycobacterium tuberculosis. They were isolated using an intriguing target-based whole-cell antisense differential sensitivity assay as inhibitors of fatty acid biosynthesis of type II. This type of biosynthesis is not present in humans. Platensimycin inhibits the elongation-condensing enzyme FabF, whereas platencin inhibits both FabF and FabH. For these antibiotics to become successful drugs, their pharmacokinetics must be improved. They have too high a rate of clearance in the body, yielding a low degree of systematic exposure. They work well when administered by continuous infusion, but this is not a useful method of delivery to patients. The two antibiotics and many analogs have been prepared by chemical synthesis. Natural congeners have also been obtained from the producing actinomycete. However, none of these molecules are as active as platensimycin and platencin. Using tools of rational metabolic engineering, superior strains have been produced making hundreds of times more antibiotic than the natural strains.
Similar content being viewed by others
Introduction
Natural products have proven to be a rich and diverse source of antibacterial leads used in the development of drugs to treat bacterial infections.1 They have provided us with the majority of antimicrobial scaffolds for many drugs. Despite the fact that natural products have revolutionized medicine, it is evident that pharmaceutical companies have significantly reduced their screening efforts for new antibiotics. It is estimated that less than 1% of prokaryotes and ∼7% of fungi have been isolated and cultured thus far. This illuminates the importance of exploring diverse geographical areas and a variety of habitats while collecting samples. Thus, there is great potential for discovering new natural products from these untapped sources. If there is any hope for a second ‘Golden Age’ of antibiotic discovery, we must put to use all of the recent advancements in bacterial genomics, screening and isolation of natural products.
Platensimycin and platencin, novel antibiotics discovered by scientists at Merck & Co., Inc.,2 represent two promising natural products.3 Not only do they show potent antibacterial activity, but they also lack any observed toxicity. Platensimycin is effective against many resistant bacteria, such as methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci.4 This compound also inhibits the highly pathogenic bacterium, Mycobacterium tuberculosis.5 Although platensimycin and platencin have some problems with regard to their pharmacokinetic properties, their mode of action is very unique. We anticipate that in the future, platensimycin and platencin, or their chemical derivatives, will be useful drugs against antibiotic-resistant Gram-positive bacterial pathogens.
Platensimycin
Platensimycin is a bacteriostatic drug with no cross-resistance to other classes of antibiotic-resistant bacteria, thereby making it a potentially useful drug candidate.4, 6 It has minimal inhibitory (MIC) values of 0.1–0.32 μg ml−1 against Gram-positive bacteria.7 Furthermore, it is relatively non-toxic to humans. Its unique target is bacterial fatty acid synthesis (FAS). In 2006, platensimycin was isolated from a soil sample collected in South Africa. It was present in one of 250 000 natural product extracts, which were screened for antibiotic properties.8, 9, 10 Merck researchers used a target-based whole-cell antisense differential sensitivity assay to identify secondary metabolites which inhibited FabH/FabF (enzymes crucial to the fatty acid biosynthetic system of bacteria).11 The idea was to lower the level of the bacterial target in the bacterial assay strain by overexpressing its antisense RNA. As a result, the genes encoding the condensing enzymes, FabF and FabH of FAS type II, would be decreased in expression. The bacterium used for the assay thus became hypersensitive to inhibitors acting on those targets.9, 10, 12
The differential sensitivity assay employed a two-plate agar diffusion technique. One plate contained an S. aureus strain with a low level of the target enzymes expressing FabF and FabH antisense RNA and another plate contained wild-type S. aureus. In essence, the researchers were searching for an extract that produced a much larger inhibition zone on the plate containing the antisense strain than on the wild-type S. aureus plate. The difference in the sizes of the zones indicated the presence of a FabF and/or a FabH inhibitor. At the molecular level, translation would be hindered in the antisense RNA strain, resulting in a reduction in the levels of the target protein.8 Hence, the decreased protein production is attributed to a lower expression of the gene product. The antisense strain contains a sequence of nucleotides, which are complementary to that of mRNA, enabling a complex to form between this strand and the naturally occurring mRNA. The formation of this complex then prevents the ribosome from readily accessing the nucleotide sequence of mRNA, which is necessary for its translation into protein. This approach was crucial for the discovery of platensimycin and platencin, which otherwise would have been missed in conventional screening assays.
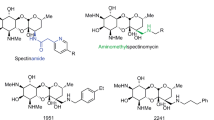
The researchers discovered that platensimycin was produced by the filamentous bacterium (actinomycete) Streptomyces platensis strain MA7327.2, 3 The antibiotic consists of two distinct structural moieties: a 3-amino-2,4-dihydroxybenzoic acid and a C-17 tetracyclic enone with an ether ring (cyclic ether, also called a tetracyclic ketolide), which are joined by an amide bond (Figure 1).13 The latter part of the molecule is hydrophobic. Platensimycin has a molecular formula of C24H27NO7 and MW of 441.47 g mol−1.
Platencin
Another antibiotic, platencin, which is actually an analog of platensimycin, also exhibits antibacterial activity against Gram-positive bacteria (MIC=0. 5–32 μg ml−1) by inhibiting FAS. In vitro studies have shown that it is effective against methicillin-resistant S. aureus, vancomycin-resistant enterococci, and both linezolid- and macrolide-resistant pathogens.10 This antibiotic represents yet another promising drug candidate due to its in vivo efficacy and absence of toxic side effects. Platencin is produced by S. platensis strain MA7339 and was discovered in a soil sample collected from Spain, in which the same whole-cell differential assay was used. Platencin has a molecular formula of C24H25NO6 and a MW of 425.47 g mol−1, slightly smaller than platensimycin. It consists of a tetracyclic unit without the ether ring (thus, different from the pentacyclic ketolide of platensimycin) that is connected by an amide bond to the aminobenzoic acid moiety (Figure 2).10, 12
Unlike platensimycin, which targets the elongation condensing enzyme FabF, platencin is a dual inhibitor of FabF and the initiation condensing enzyme FabH.10, 12 The fact that platencin essentially targets two enzymes in the FAS pathway may contribute to a decrease in the potential for resistance development against the drug.
Biosynthetic studies
An understanding of the biosynthetic pathways of platensimycin and platencin is important for improving the titer of these compounds and developing new natural analogs. The biosynthesis of these novel antibiotics was studied via incorporation of stable-isotope precursors.14, 15 Different isotopes of labeled sodium acetate and sodium pyruvate were fed to the producing organisms to identify the enrichment of specific carbon atoms in each of the molecules. Both sodium acetate and sodium pyruvate showed incorporation into the 3-amino-2,4-dihydroxybenzoic acid moiety of platencin and platensimycin. The aminobenzoic acid moiety was found to be synthesized from pyruvate and acetate via the tricarboxylic acid cycle and phosphoenolpyruvate. However, the C-17 polycyclic enone acid moiety incorporated pyruvate and was biosynthesized by a non-mevalonate terpenoid pathway, often found in eubacteria, green algae, actinomycetes and higher plants.14, 15, 16
Chemical synthesis of platensimycin and platencin
McGrath et al.17 explored how the center of the pentacyclic ketolide of platensimycin was formed. Structural studies revealed that the compound consisted of a compact core that was connected through a propionate tether to a highly oxygenated aromatic ring. Through the use of a copper-catalyzed oxirane ring expansion and an alkylative dearomatization, they developed a concise synthetic route to the platensimycin core. Other important aspects of their approach were phenol ether deprotection, nucleophilic enoate epoxidation and the introduction of a substituted alkyl ketone, using a trifluoroborate cross coupling.
Nicolaou et al.18 developed an original chemical synthetic approach for platencin. As part of their synthesis, they used a cobalt-catalyzed asymmetric Diels–Alder reaction and a one-pot reductive rearrangement of bicyclic ketones to bicyclic olefins. Through their efforts, they were able to prepare both the racemic (±) and naturally occurring (−) forms of platencin. This was important, as it allowed researchers to perform the first X-ray crystallographic analysis of this natural product.
Despite many of the successful formal syntheses of platensimycin and platencin that have been proposed, Tiefenbacher and Mulzer19 sought to develop a more concise total synthesis of natural (−) platencin. Starting with perillaldehyde, they obtained 32 mg of platencin in nine linear steps. Some of the essential features included a highly diastereoselective Diels–Alder reaction with the reactive Rawal's diene (leading to the formation of an all-carbon quaternary center) and a ring-closing metathesis, which generated a tricyclic skeleton. In addition, they also utilized a hydration/dehydration technique to convert the endocyclic alkene to an exocyclic alkene (the latter of which was referred to as the exo-olefin). Another notable aspect of their short synthesis was the 1,4-addition of a ketone enolate to methyl acrylate. This created the second all-carbon quaternary center. With an overall yield of 10%, their synthesis of platencin was comparable to previous ones.
Mode of action
Most antibiotics work by blocking the production of proteins, DNA or the bacterial cell wall.20 However, platensimycin is unique in the sense that it does not inhibit DNA, RNA, protein or cell wall biosynthesis.9 This contributes to a lower probability of bacterial resistance. Instead, platensimycin exerts its antibacterial activity by targeting type II FAS (FASII). It is important to differentiate between the fatty acid synthetic system of prokaryotes such as bacteria, plants and protozoa (type II) from that found in eukaryotes (mammals and fungi; type I).21 In type I FAS (the associated system), a multifunctional polypeptide with several domains carries out all the steps in the biosynthetic pathway. On the other hand, type II FAS (dissociated system) consists of many discrete enzymes, each of which has an integral role in a single step of FAS. These enzymes are involved in condensation, reduction and dehydration.22 The fact that the FASII system is present in bacteria, but not in humans, explains why it is such a good target for antibacterial drug discovery.8 In addition, the FabF enzyme is highly conserved across many bacterial genomes, implying a potential for broad-spectrum antibiotic activity in the future.
The synthesis of fatty acids is essential for bacterial viability, as they make up the cell membranes of bacteria.8, 12 These fatty acids form the building blocks of many cellular structures and are required for energy storage. Platensimycin interferes with this process by selectively inhibiting the β-ketoacyl (acyl-carrier protein) ACP synthase (FabF enzyme). FabF is an elongation-condensing enzyme whose main function is to add acetate units to the growing fatty acid chain. Platensimycin must compete with malonyl-ACP (the normal substrate) for the malonate-binding site of FabF. In doing so, this enables platensimycin to interact with the acyl-enzyme intermediate of FabF and thereby inhibit FAS.2 Researchers used a radioactively labeled derivative of platensimycin (3H-dihydroplatensimycin) to confirm that the drug selectively targets the acyl intermediate of FabF.6, 9
In a direct comparison of platensimycin and platencin using a gel elongation assay,10 the IC50 of platensimycin against FabF was 0.29 μM. It was very poorly active against FabH, with an IC50 of 247 μM. Platencin was less active than platensimycin against FabF with an IC50 of 4.58 μM, but was more active than platensimycin against FabH (IC50 of 9.17 μM). Thus, platensimycin showed a 15-fold superiority over platencin against FabF. On the other hand, platencin exhibited a 27-fold higher activity over platensimycin against FabH.7
X-ray crystallographic studies have revealed that the formation of the acyl-FabF intermediate further exposes the enzyme's active site by allowing ACP to be released.9, 22 The acyl-enzyme intermediate must form before platensimycin can interact with the malonyl-binding site of FabF. The researchers were unable to stabilize this intermediate within the active site of the wild-type FabF enzyme.9 Therefore, the group used an Escherichia coli FabF-mutant strain (C163Q), which showed a 50-fold increase in the binding affinity of platensimycin. A specific conformational change, which is known as the ‘open configuration’, is required for platensimycin to bind the enzyme.6 Once bound to the active site of FabF, platensimycin engages in hydrogen-bonding interactions with key amino acid residues such as alanine 309, histidine 303, histidine 340 and threonine 270.9 The ability of platensimycin to bind to the enzyme and form hydrogen bonds is highly dependent upon the conformation of the cyclohexenone ring.2 This, in turn, affects the biological activity of platensimycin by halting the elongation of the fatty acid chain, eventually leading to inhibition of the fatty acid synthetic pathway.
Platensimycin is not effective against Gram-negative pathogens, due, in part, to efflux mechanisms, which enable these bacteria to pump certain compounds (antibiotics) out of their cells. When such pumps are disabled due to mutation, such as that seen in the efflux-negative E. coli (tolC), platensimycin is capable of inhibiting Gram-negative bacteria as well.4, 9, 22 Perhaps an even better alternative for treating Gram-negative bacterial infections would be administering platensimycin along with a second drug that could block the efflux pumps.4 Of great interest is the ability of platensimycin to inhibit M. tuberculosis; it also inhibits M. smegmatis.5 Specifically inhibited was the biosynthesis of mycolic acid. Interestingly, platensimycin did not inhibit the β-ketoacyl-ACP synthase FabH in M. tuberculosis. Rather, it inhibited KasA and KasB, two β-ketoacyl-ACP synthases involved in mycolic acid formation, which is essential for mycobacterial survival.
Pharmacokinetics
Although platensimycin and platencin provide a solution to fighting antibiotic-resistant bacteria, the pharmacokinetics of these two natural products has prevented them from entering clinical trials. Pharmacokinetics, which explores the fate of a drug inside the body, is important for such a new chemical class of antibiotics, because it allows researchers to determine if they are safe and/or effective for human use. Indeed, it has complicated the process of turning them into commercial drugs.20 The research team at Merck believes that platensimycin and platencin have a high rate of clearance, resulting in a low degree of systematic exposure; therefore, they need to be administered i.v. with a pump.3, 9 Experimentation with mice confirmed that the continuous infusion of platensimycin into the cells led to the elimination of an S. aureus infection.9, 20 However, continuous infusion is not an optimal route of delivery of an antibiotic, as it is not desirable for patients, thus indicating platensimycin's poor pharmacokinetics.22 They also found that these compounds are less potent when given either s.c. or orally. In other words, their efficacy is greatly reduced when given through more conventional routes.14 This suboptimal delivery system emphasizes the need for modification of antibiotic structure.23
The search for analogs/derivatives of platensimycin and platencin
The exploration of the FASII pathway for potential drug targets had begun well before the emergence of platensimycin and platencin. Although no drugs targeting the condensing enzymes are currently used in modern medicine, there was the discovery of two natural products, cerulenin and thiolactomycin, more than 20 years ago.24, 25, 26 Similar to platensimycin, cerulenin selectively inhibits the elongation-condensing enzyme, FabF. On the other hand, thiolactomycin, like platencin, is actually a dual inhibitor of FabH and FabF. However, both of these FASII inhibitors exhibit poor antibacterial activity, which may be attributed to the fact that these compounds have trouble reaching their intracellular targets.12 Despite these difficulties, researchers at Merck used the antisense technology along with both a gel elongation assay and whole-cell-based two-plate assay to discover the following inhibitors of FabF/B and FabH: phomallenic acids A, B and C.8 These new inhibitors exhibited good antibacterial activity against S. aureus, Haemophilus influenzae, Bacillus subtilis and E. coli. Of all three compounds, phomallenic acid C showed the best antibacterial activity; it was 20-fold better than cerulenin and thiolactomycin against S. aureus. However, because of toxicity, it was never developed into a drug.
As mentioned above, poor pharmacokinetics have prevented platensimycin and platencin from entering clinical trials. However, an understanding of the structural elements, which are essential for antibiotic activity, may provide a foundation for the development of analogs of these compounds. Thus, the pharmacokinetic problem associated with platensimycin and platencin might be overcome through chemical modification and the development of new congeners.27
In an attempt to improve the pharmacokinetic properties of platensimycin, researchers at Merck focused on chemically modifying its structure.13 When platensimycin was treated with diazomethane at controlled temperature, the phenolic groups of this compound were selectively methylated. Some additional experiments they conducted were halogenation, reduction, epoxidation and Bayer–Villiger oxidation, each of which resulted in different functional groups added to platensimycin. They reported that the catalytic hydrogenation of platensimycin produced dihydroplatensimycin, which was then converted to a cyclic enamino-amido product. It is important to note that platensimycin showed better activity than all of the compounds formed from these reactions.
Merck scientists also focused their efforts on modifying the enone moiety, based on studies of the structure–activity relationship of platensimycin.28 They discovered a total of nine analogs, which inhibited FabF and thus exhibited antibacterial activity. Three different assays were used to conduct these studies: 1) whole-cell sa (S. aureus) assay, 2) inhibition of FabF in a cell-free FASII assay and 3) antisense FabH/FabF, in which S. aureus was sensitized (referred to as saFabF2). The main objective was to determine whether the enone moiety was critical for the biological activity of platensimycin. Of all nine analogs, compounds 2d and 2g, which possessed the ketone moiety and similar conformation, were most active in the saFabF2 antisense assay. The minimum detection concentration for compound 2d was 0.1 μg ml−1 and that of 2g was 0.03 μg ml−1. However, none of the analogs exhibited better antibacterial activity than platensimycin. Another nine analogs were isolated from S. platensis; that is, compounds 11–20.29 These were hydroxylated congeners of platensimycin and platencin, and methyl esters presumably derived by cytochrome P450 oxidation of the terpenoid units post cyclization. However, they were all less active than the two antibiotics.
Nicolaou et al.18, 30, 31, 32, 33 reported several strategies for the total synthesis of platensimycin and related natural products. By using platensimycin/platencin as a starting point, they explored the relationship between structure and function. They first began by constructing the cage-like core of platensimycin and then added various side chains through amide bond formation. This was done simply to ensure that the syntheses for platensimycin and its congeners were reliable. They developed two asymmetric routes to this core structure. For the first, a rhodium-catalyzed asymmetric cycloisomerization involving a terminal acetylene was utilized as a general method for obtaining terminal enynes. The second route used a hypervalent iodine-mediated de-aromatizing cyclization of an enantiopure substrate. A samarium diiodide-mediated ketyl radical cyclization and an acid-catalyzed etherification were employed to form the last two bonds of the cage-like core.
In altering the structure of certain parts of the platensimycin molecule, primarily the tetracyclic enone core, the Nicolaou group studied how this would affect its biological profile. Two of the derivatives, which they synthesized de novo, were adamantaplatensimycin and carbaplatensimycin. Both of these new analogs showed strong antibacterial activity against Gram-positive pathogens such as methicillin-resistant S. aureus and vancomycin-resistant enterococci; however, they were not as potent as platensimycin. The relationship between structure and function of these compounds must be further studied to develop new analogs with even better antibiotic activity and pharmacokinetics. These synthetic approaches could greatly facilitate the development of new natural analogs of platensimycin.
Another way to improve the pharmacokinetics of platensimycin is through the discovery of natural congeners from fermentation broths.34 Merck researchers discovered three new congeners of platensimycin: methylplatensinoate, platensic acid and platensimide A from the fermentation broth by chemical screening. Using a FASII assay in a cell-free system, methylplatensinoate (denoted as compound 3) inhibited S. aureus with an IC50=167 μg ml−1. The IC50 of platensic acid was >333 μg ml−1. Platensimide A showed the best antibacterial activity, with an IC50=80 μg ml−1. None of the three compounds inhibited S. aureus, Streptococcus pneumoniae or Enterococcus faecalis at 64 μg ml−1. In the antisense two-plate differential sensitivity assay, platensimide A exhibited a minimum detection concentration of 100 μg ml−1, which was 2500-fold lower activity than that of platensimycin. A minimum detection concentration is the minimum concentration of the compound that is required to differentiate between the zones of inhibition on the antisense and control plate.34 Platensimide A showed the best activity of all the compounds, yet none of the three congeners had an MIC <64 μg ml−1. In comparison, the MIC of platensimycin was 0.5 μg ml−1 for the same S. aureus strain. The data clearly indicate that because these compounds lack the 3-amino-2,4-dihydroxybenozoic acid moiety (which is characteristic of both platensimycin and platencin), they have poorer antibacterial activity. In 2008, Merck researchers reported the discovery of two more natural congeners of platensimycin from the fermentation broth.16 They isolated homoplatensimide A (compound 3a) and its methyl ester (compound 3b), which were very important in helping to explain the biosynthetic origin of platensimycin's C-17 tetracyclic enone. Earlier studies had shown that this moiety (known as platensic acid) was derived from a non-mevalonate terpenoid pathway.14, 15 Homoplatensimide A actually contained a C-20 diterpenoid tetracyclic enoic acid that was coupled with glutamine.16 This C-20 diterpenoid, referred to as homoplatensic acid, was very likely the biosynthetic source of the C-17 part of platensimycin. In the FASII assay, homoplatensimide A poorly inhibited S. aureus, with an IC50 of >167 μg ml−1 (again, likely due to the lack of the aminobenzoic acid moiety). For the antisense two-plate differential sensitivity assay, this compound exhibited a minimum detection concentration of 400 μg ml−1, which was about 10 000-fold lower activity than that of platensimycin.
The Merck research group in 2009 reported the discovery of four new congeners of platensimycin: (i) platensimycin A1 (ii) its methyl ester, (iii) hydroxyplatensic acid and (iv) its methyl ester. Platensimycin A1 and its methyl ester closely resemble platensimycin, except for the substitution of a hydroxyl group at C-14.35 This addition of –OH at C-14 actually decreased the activity of the first two compounds. On the other hand, the last two congeners, hydroxyplatensic acid and its methyl ester, both lack the benzoic acid moiety characteristic of platensimycin. However, they still possess the tetracyclic enone portion of the molecule along with a substituted hydroxyl group. In 2010, they isolated three hydroxylated analogs of platencin; that is, platencins A2, A3 and A4 from the broth of S. platensis MA7327.36 These analogs were hydroxylated in the terpenoid portion of the molecule and were much less active than platencin.
Genetic engineering of S. platensis strains for titer improvement
Improving fermentation titers of new drugs derived from natural products is essential to the commercialization process. For many years and still to this day, the traditional method used in industry for strain improvement has been ‘random mutation and screening’. Yet, one of the reasons why this approach is not the most efficient is the significant amount of time and effort required. An alternative strategy for titer improvement is ‘metabolic engineering’, which often involves the manipulation of pathway-specific regulatory elements that govern the production of secondary metabolites. Researchers have been able to show that improvement in production of secondary metabolites by Streptomyces could occur in both native producers and heterologous hosts. The advancements in the fields of molecular microbiology and recombinant DNA technology have greatly facilitated the cloning and sequencing of secondary metabolite gene clusters. This, in turn, has led to the identification and characterization of many pathway-specific regulators. To utilize the strategy of metabolic engineering, the difference between manipulating positive and negative regulators must be understood. By overexpressing the pathway-specific activator genes, a dramatic increase in titer was observed in engineered strains, such as the producers of fredericamycin and C-1027.37 On the other hand, in negative regulation, pathway-specific repressors could be deleted (by gene knockout), which would have the same effect of increasing secondary metabolite titers.
The genetic manipulation of the fatty acid biosynthetic pathway is essential in improving the titers of drug candidates such as platensimycin and platencin. Shen's group at the University of Wisconsin used PCR to identify and clone the specific locus of the biosynthetic genes responsible for the production of these compounds, including ptmR1.37, 38 This gene is a negative regulator of transcription. A fragment of the 3-amino-5-hydroxybenzoic (AHBA) synthase gene, which was believed to have a role in the biosynthesis of the benzoic acid moiety of platensimycin, was amplified using this PCR-based approach. They found that deletion or inactivation of ptmR1 in strain MA 7339 greatly altered the production of platensimycin.23 The resulting over-producing strain was S. platensis SB12002, with a platensimycin titer of 323 mg l−1, representing about a 100-fold greater titer than the original strain.
Similar studies were done with the platencin producer S. platensis MA7339.38 Inactivation of the gene ptnR1, a homolog of ptmR1, resulted in about a 100-fold increase in the production of platencin.37 The new strain, S. platensis SB12600, produced 255 mg l−1 of platencin. Interestingly, the new strain also produced platencin A1 and eight new congeners, platencins A2–A9. Compound 5 (platencin A3) exhibited the best antibacterial activity of all the congeners, with an MIC of 8 μg ml−1 against a strain of methicillin-resistant S. aureus. Such improvements in titer should aid in the development of these compounds into clinically useful antibacterial agents.
Final comments
Bacterial fatty-acid synthesis appears to be a novel antibacterial target. It has enabled scientists to thoroughly analyze enzymes integral to FAS to determine if they are suitable candidates for fighting bacterial infections. The viability of bacteria is entirely dependent on this pathway, because fatty acids are the main component of their cell membranes. As described above, the discovery of platensimycin and platencin resulted from an ingenious assay procedure. Also impressive was the marked increase (∼100-fold) in the production of these compounds by application of rational metabolic engineering. More research needs to be devoted to the exploration of these regulatory pathways to continue improving production of these compounds. Nothing has been published on the effect of media manipulation on the biosynthesis of these compounds. There is much that has to be learned about the effects of primary metabolite addition to media, as well as investigation of regulatory controls on their production. However, the greatest need for future development of platensimycin and platencin is the improvement in their pharmacokinetic properties.
References
Singh, S. B. & Barrett, J. F. Empirical antibacterial drug discovery-foundation in natural products. Biochem. Pharmacol. 71, 1006–1015 (2006).
Singh, S. et al. Isolation, structure, and absolute stereochemistry of platensimycin, a broad spectrum antibiotic discovered using an antisense differential screening strategy. J. Am. Chem. Soc. 128, 11916–11920 (2006).
Arnaud, C. Antibiotic halts lipid synthesis. Chem. Eng. News 84, 7 (2006).
Potera, C. Novel pentacyclic from soils blocks bacterial fatty acid synthesis. Microbe 1, 350–351 (2006).
Brown, A. K., Taylor, R. C., Bhatt, A., Fuetterer, K. & Besra, K. Platensimycin activity against mycobacterial β-ketoacyl ACP synthases. PLOS One 4, eb306 (2009).
Barton, S. New antibiotic on the horizon? Nat. Rev. Microbiol. 4, 571 (2006).
Singh, S. B. & Young, K. New antibiotic structures from fermentations. Expert Opin. Ther. Pat. 20, 1359–1371 (2010).
Young, K. et al. Discovery of FabH/FabF inhibitors from natural products. Antimicrob. Ag. Chemother. 50, 519–526 (2006).
Wang, J. et al. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature 441, 358–361 (2006).
Wang, J. et al. Discovery of platencin, a dual FabF and FabH inhibitor with in vivo antibiotic properties. Proc. Natl. Acad. Sci. USA 104, 7612–7616 (2007).
Singh, S. B., Phillips, J. W. & Wang, J. Highly sensitive target-based whole-cell antibacterial discovery strategy by antisense RNA silencing. Curr. Opin. Drug Discov. Devel. 10, 160–166 (2007a).
Jayasuriya, H. et al. Isolation and structure of platencin: A FabH and FabF dual inhibitor with potent broad-spectrum antibiotic activity. Angew. Chem. Int. Ed. Engl. 46, 4684–4688 (2007).
Singh, S. B., Herath, K. B., Wang, J., Tsou, N. & Ball, R. G. Chemistry of platensimycin. Tetrahedron Lett. 48, 5429–5433 (2007b).
Herath, K. B, Attygalle, A. B. & Singh, S. B Biosynthetic studies of platensimycin. J. Am. Chem. Soc. 129, 15422–15423 (2007).
Herath, K. B., Attygalle, A. B. & Singh, S. B. Biosynthetic studies of platencin. Tetrahedron Lett. 49, 5755–5758 (2008a).
Jayasuriya, H. et al. Structure of homoplatensimide A: a potential key biosynthetic intermediate of platensimycin isolated from Streptomyces platensis. Tetrahedron Lett. 49, 3648–3651 (2008).
McGrath, N. A., Bartlett, E. S., Sittihan, S. & Njardarson, J. T. A concise ring-expansion route to the compact core of platensimycin. Angew. Chem. Int. Ed. Engl. 48, 8543–8546 (2009).
Nicolaou, K. C., Tria, G. S., Edmonds, D. J. & Kar, M. Total syntheses of (±)-platencin and (-)-platencin. J. Am. Chem. Soc. 131, 15909–15917 (2009a).
Tiefenbacher, K. & Mulzer, J. A nine-step total synthesis of (-)-platencin. J. Org. Chem. 74, 2937–2941 (2009).
Pearson, H. Antibiotic faces uncertain future. Nature 441, 260–261 (2006).
Payne, D. J., Warren, P. V., Holmes, D. J., Ji, Y. & Lonsdale, J. T. Bacterial fatty-acid biosynthesis: a genomics-driven target for antibacterial drug discovery. Drug Disc. Today 6, 537–544 (2001).
Manallack, D. T., Crosby, I. T., Khakham, Y. & Capuano, B. Platensimycin: a promising antimicrobial targeting fatty acid synthesis. Curr. Med. Chem. 15, 705–710 (2008).
Smanski, M. J., Peterson, R. M., Rajski, S. R. & Shen, B. Engineered Streptomyces platensis strains that overproduce antibiotics platensimycin and platencin. Antimicrob. Ag. Chemother. 53, 1299–1304 (2009).
Matsumae, A., Nomura, S. & Hata, T. Studies on cerulenin. IV. Biological characteristics of cerulenin. J. Antibiot. 17, 1–7 (1964).
Noto, T., Miyakawa, S., Oishi, H., Endo, H. & Okazaki, H. Thiolactomycin, a new antibiotic. III. In vitro antibacterial activity. J. Antibiot. 35, 401–410 (1982).
Omura, S. et al. Thiotetromycin, a new antibiotic. Taxonomy, production, isolation, and physicochemical and biological properties. J. Antibiot. 36, 109–114 (1983).
Lu, X. & You, Q. Recent advances on platensimycin: a potential antimicrobial agent. Curr. Med. Chem. 17, 1139–1155 (2010).
Shen, H. C. et al. Synthesis and biological evaluation of platensimycin analogs. Bioorg. Med. Chem. Lett. 19, 1623–1627 (2009).
Zhang, C. et al. Platensimycin and platencin congeners from Streptomyces platensis. J. Nat. Prods. 74, 329–340 (2011).
Nicolaou, K. C., Lister, T., Denton, R. M., Montero, A. & Edmonds, D. J. Adamantaplatensimycin a bioactive analogue of platensimycin. Angew. Chem. Int. Ed. Engl. 46, 4712–4714 (2007a).
Nicolaou, K. C. et al. Total synthesis and antibacterial properties of carbaplatensimycin. J. Am. Chem. Soc. 129, 14850–14851 (2007b).
Nicolaou, K. C. et al. Design, synthesis and biological evaluation of platensimycin analogues with varying degrees of molecular complexity. J. Am. Chem. Soc. 130, 13110–13119 (2008).
Nicolaou, K. C., Li, A., Edmonds, D. J., Tria, G. S. & Ellery, S. P. Total synthesis of platensimycin and related natural products. J. Am. Chem. Soc. 131, 16905–16918 (2009b).
Herath, K. B. et al. Structure and semisynthesis of platensimide A, produced by Streptomyces platensis. Org. Lett. 10, 1699–1702 (2008b).
Singh, S. B. et al. Isolation, enzyme-bound structure, and activity of platensimycin A1 from Streptomyces platensis. Tetrahedron Lett. 50, 5182–5185 (2009).
Zhang, C. et al. Isolation, structure and biological activities of platencin A2–A4 from Streptomyces platensis. Bioorg. Med. Chem. 18, 2602–2610 (2010).
Chen, Y., Smanski, M. J. & Shen, B. Improvement of secondary metabolite production in Streptomyces by manipulating pathway regulation. Appl. Microbiol. Biotechnol. 86, 19–25 (2010).
Yu, Z. et al. Engineering of Streptomyces platensis MA7339 for overproduction of platencin and congeners. Org. Lett. 12, 1744–1747 (2010).
Acknowledgements
We thank Dr. Sheo Singh of Merck & Co., Inc. for reading the manuscript and suggesting important modifications. We also acknowledge the encouragement by Drs. Vincent Gullo and Jon Kettenring of R.I.S.E.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Martens, E., Demain, A. Platensimycin and platencin: promising antibiotics for future application in human medicine. J Antibiot 64, 705–710 (2011). https://doi.org/10.1038/ja.2011.80
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2011.80
Keywords
This article is cited by
-
Nocardia noduli sp. nov., a novel actinobacterium with biotechnological potential
Archives of Microbiology (2022)
-
Highly reproducible solid-phase extraction membrane for removal and surface-enhanced Raman scattering detection of antibiotics
Journal of Materials Science (2018)
-
Nutritional control of antibiotic production by Streptomyces platensis MA7327: importance of l-aspartic acid
The Journal of Antibiotics (2017)
-
Titer improvement and pilot-scale production of platensimycin from Streptomyces platensis SB12026
Journal of Industrial Microbiology and Biotechnology (2016)
-
The re-emergence of natural products for drug discovery in the genomics era
Nature Reviews Drug Discovery (2015)