Abstract
A Gram-positive aerobic actinomycete, designated SR14.14T, isolated from the rhizospheric soil of rubber tree was determined taxonomically using a polyphasic approach. The organism contained meso-diaminopimelic acid and the N-acetyl type of peptidoglycan. The predominant menaquinones were MK-9, MK-9(H2) and MK-9(H4). Madurose was detected in the whole-cell hydrolysates. Mycolic acids were not presented. Major phospholipids were diphosphatidylglycerol, phosphatidylethanolamine and phosphatidylinositol mannoside. Major cellular fatty acid was iso-C16: 0 and the G+C content was 71.9 mol%. Phylogenetic analysis based on 16S rRNA gene sequence suggested that the isolate belongs to the genus Sphaerisporangium. The sequence similarity value between the strain SR14.14T and its closely related species, Sphaerisporangium album, was 97.8%. DNA–DNA hybridization values between them were well below 70%. Based on genotypic and phenotypic data, strain SR14.14T represents a novel species in the genus Sphaerisporangium, for which the name Sphaerisporangium siamense sp. nov. is proposed. The type strain is SR14.14T (=BCC 41491T=NRRL B-24805T=NBRC 107570T).
Similar content being viewed by others
Introduction
The genus Sphaerisporangium was proposed by Ara and Kudo1 with Sphaerisporangium melleum as type species. This genus was emended by Cao et al.2 for the strain containing MK-9(H6), MK-9(H4), MK-9(H2) and MK-9 as predominant menaquinones and the DNA G+C contents of 67–72 mol%. The genus formed a phyletic line within the 16S rRNA genes tree of the Streptosporangiaceae, the family of which included the following genera: Acrocarpospora, Herbidospora, Microbispora, Microtetraspora, Nonomuraea, Planobispora, Planomonospora, Planotetraspora, Sphaerisporangium, Streptosporangium and Thermopolyspora.1, 3, 4 At the time of writing, the genus Sphaerisporangium encompassed six species. Sphaerisporangium album and S. flaviroseum were isolated from forest soil samples in China.2 Sphaerisporangium cinnabarinum, S. melleum and S. rubeum were isolated from sandy soil samples in Bangladesh.1 Sphaerisporangium viridialbum was transferred from Streptosporangium viridialbum.1, 5
During our investigation of actinomycetes from rhizospheric soil of rubber tree collected from Phatthalung province, Thailand, the strain SR14.14T was isolated. A polyphasic study was designed to establish the taxonomic status of this strain. Resultant data based on this polyphasic study showed that this isolate represented a novel species of the genus Sphaerisporangium.
Materials and methods
Strain SR14.14T was isolated from the rhizospheric soil of rubber tree using the spread plate method on starch casein agar,6 supplemented with ketokonazole (100 μg ml−1) and nalidixic acid (25 μg ml−1). The plates were incubated at 28 °C for 2 weeks. The strain was purified and maintained on glucose yeast extract (GYE) agar (containing glucose 1.0% (w/v), yeast extract 1.0% (w/v) and agar 1.5% (w/v)) at room temperature. Suspensions of spores or mycelium were stored at −20 °C in 20% glycerol and lyophilized for long-term preservation. Sphaerisporangium album DSM 45172T was used in this study for comparison purposes.
Cultural characteristics were determined using 14-days culture on ISP media 2, 3, 4 and 5 of Shirling and Gottlieb7 (Difco, Detroit, MI, USA) oatmeal-nitrate agar (JCM medium 52; Difco) and yeast-starch agar (JCM medium 42; Difco) at 27 °C. The color of mycelium and soluble pigment were determined by comparison with the color of chips in the Color Harmony Manual.8 The morphological characteristics were observed by light microscopy and scanning electron microscopy (JEOL-JSM 5600 LV, Japan) of 14-day-old cultures grown on ISP medium 3. Motility was observed with a light microscope using cells grown on ISP medium 2 for 14 days and incubated in soil-extracted solution (5 g garden soil in 25 ml distilled water, mixed well and filtered through a 0.22-μm Millipore membrane (Millipore, Darmstadt, Germany)) for 30 min.
The utilization of a variety of substrates as sole carbon sources was tested using the basal inorganic nitrogen medium9 supplemented with a final concentration of 1% (v/v) of the filter sterile-tested carbon sources. Catalase and oxidase activities were determined with 3% (v/v) hydrogen peroxide solution and 1% tetramethyl p-phenylenediamine dihydrochloride solution, respectively. Growth at various pH values and the tolerance of NaCl were examined on ISP medium 2. The temperature range for growth was determined on ISP medium 2 using a temperature gradient incubator (Tokyo Kagaku Sangyo, Tokyo, Japan) between 5 and 50 °C. Enzyme activity profiles were carried out using the API ZYM (bioMèrieux) test kits. Melanin pigment was examined on ISP medium 7.7 The production of hydrogen sulfide was detected using lead acetate strips. Hydrolysis of casein, gelatin and nitrate reduction was examined by following the methods of Gordon and Mihm.10
Biomass used for chemotaxonomic analyses was obtained from cultures grown in shake flasks of ISP medium 2 for 10 days at 27 °C. The cells were harvested by centrifugation and washed three times with distilled water before freeze-drying. Standard procedures were used to determine the isomers of diaminopimelic acid.11, 12 The acyl type of the cell wall was analyzed according to the method of Uchida and Aida.13 Whole-cell sugars were analyzed according to the method of Becker et al.11 Polar lipids were examined using two-dimensional TLC and identified by the method of Minnikin et al.14 Menaquinones were extracted and purified by the method of Collins et al.,15 and isoprene units were subsequently analyzed by LC/MS (JMS-T100LP, JEOL) with a PEGASIL ODS column (2ø × 50 mm; Senshu, Tokyo, Japan) using methanol/2-propanol (7:3). Mycolic acids were detected by TLC according to the method of Tomiyasu.16 The G+C content (mol%) of the DNA was determined by HPLC according to the method of Tamaoka and Komagata.17 The cellular fatty acid composition was analyzed by TechnoSuruga (Shizuoka, Japan) according to the instructions of the Microbial Identification System (MIDI, version 4.5) using a gas chromatograph (model HP6890; Hewlett Packard, Palo Alto, CA, USA) and identified with the ACTIN1 database. DNA–DNA relatedness was measured fluorometrically using the microplate hybridization method.18
The phylogenetic position of the isolate SR14.14T was determined based on 16S rRNA gene sequence. Genomic DNA was isolated and purified from biomass following the method of Kieser et al.19 The 16S rRNA gene was amplified as described by Duangmal et al.20 The PCR products were sequenced (First Base, Seri Kembangan, Malaysia) using universal primers.21 The resultant 16S rRNA gene sequence was aligned with the corresponding sequences of representatives of the genus Sphaerisporangium and related genera, retrieved from the GenBank databases, using the CLUSTAL X22 and PHYDIT programs (http://plaza.snu.ac.kr/~jchun/phydit/). Phylogenetic trees were inferred by the least-squares,23 maximum-parsimony24 and neighbor-joining25 tree-making algorithms from the PHYLIP suite of programs26 and TREECON software.27 The resultant phylogenetic trees were viewed by TREEVIEW program.28
Results and discussion
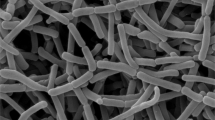
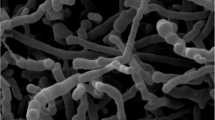
Strain SR14.14T exhibited phenotypic properties typical of members of the genus Sphaerisporangium. The strain was Gram-positive, non-acid–alcohol-fast, and produced branched and non-fragmented substrate mycelia. Spherical spore vesicles were born on aerial mycelia with non-motile spores. Observations using scanning electron microscopy indicated that the sizes of spherical spore vesicles were between 4–5 μm (Figure 1). Good growth of pale pink to pale orange substrate mycelia was observed on ISP medium 2, oatmeal-nitrate agar and yeast-starch agar. The strain formed white aerial hyphae with moderate sporulation on ISP medium 2, ISP medium 3, oatmeal-nitrate agar and yeast-starch agar. Light yellow soluble pigment was produced on ISP medium 3 and oatmeal-nitrate agar. Poor growth was observed on ISP media 4 and 5. Melanin pigment was not observed.
The results from chemical analysis indicated that strain SR14.14T has chemotaxonomic markers typical of the genus Sphaerisporangium. The strain contained meso-diaminopimelic acid in the peptidoglycan. The predominant menaquinones were MK-9, MK-9(H2) and MK-9(H4). Galactose, glucose, madurose, mannose and ribose were detected as the major sugars in the whole-cell hydrolysates. Polar lipid analysis showed that the organism contained diphosphatidylglycerol, phosphatidylethanolamine and phosphatidylinositol mannoside as major phospholipids. Small amounts of phosphatidylinositol and phosphoglycolipid were also detected. Similar observations of phosphatidylinositol and phosphoglycolipid were reported by Cao et al.2 for S. album and S. flaviroseum. The cellular fatty acid profile was represented by the predominance of iso-C16: 0 (47.9%). Fatty acids found in smaller proportions included 10methyl-C17: 0 (14.7%), 10methyl-C16: 0 (8.2%), iso G-C16: 1 (4.0%), C16: 0 (3.6%), 10methyl-C18: 0 (3.2%), C18: 0 (2.5%), C17: 0 (2.3%), anteiso-C17: 0 (2.3%), C15: 0 (2.0%), iso-C15: 0 (1.8%), iso-C18: 0 (1.7%), cis 9-C16: 1 (1.1%), iso-C14: 0 (1.0%), iso-C17: 0 (1.0%), anteiso-C15: 0 (0.6%) and cis9-C17: 1 (0.6%). No mycolic acids were detected. The G+C content was 71.9 mol%.
The almost-complete 16S rRNA gene sequence (1419 nt) of strain SR14.14T was compared against related sequences in the GenBank database. The results indicated that this isolate belongs to the genus Sphaerisporangium. The strain SR14.14T showed the highest level of 16S rRNA gene similarity with the type strain of S. album (97.8%, a value corresponding to 29 nucleotide differences from 1328 locations). The 16S rRNA gene sequence similarity value of strain SR14.14T with the type strains of S. melleum, S. viridialbum and S. cinnabarinum were 97.8, 97.6 and 97.2%, respectively.
It is evident from the phylogenetic tree with members of the genus Sphaerisporangium and type strains representing species of the family Streptosporangiaceae that strain SR14.14T is closely related to S. album, with a bootstrap value of 75% by neighbor-joining analysis (Figure 2). Similar tree topographies were obtained when the least-squares and maximum-parsimony methods were applied. The isolate showed low bootstrap values with other members of the genus Sphaerisporangium. DNA–DNA relatedness studies were carried out between strain SR14.14T and S. album DSM 45172T by reciprocal hybridizations. The DNA–DNA reassociation values were in the range of 32–39%, which confirmed that the strain SR14.14T represented a separate genomic species based on whole-genomic DNA relatedness of less than the 70% cut-off point.29
Neighbor-joining phylogenetic tree based on nearly complete 16S rRNA gene sequences showing the position of the strain SR14.14T in the genus Sphaerisporangium. Asterisks indicate branches that were also found using the least-squares23 and maximum-parsimony24 tree-making algorithms. Numbers at the nodes indicate the percentage bootstrap support from an analysis of 1000 resampled datasets (only values greater than 50% are shown). Bar, 0.02 substitutions per site.
Strain SR14.14T was positive for catalase and oxidase. The temperature range for growth was 16–40 °C and good growth was observed at 26–35 °C. No growth was observed above 40 °C. The strain was able to grow at pH 5.0–9.0 and 0–2% NaCl. The strain was readily differentiated from its closest relative, S. album DSM 45172T, on the basis of physiological properties (Table 1), in particular aesculin, arbutin and xylan hydrolysis, which were positive in S. album but negative in strain SR14.14T. The utilization of arabinose, D(−)fructose, myo-inositol, D(−)mannitol, melibiose, L(−)rhamnose and sucrose was negative in SR14.14T, properties that were positive in S. album. Moreover, the strain SR14.14T produced a light yellow pigment on ISP medium 3 and oatmeal-nitrate agar after incubation at 27 °C for 2 weeks, but none was detected in S. album.
It is evident on the basis of genotypic and phenotypic data that strain SR14.14T can be distinguished from members of the genus Sphaerisporangium. We concluded that strain SR14.14T represented a novel species of Sphaerisporangium, for which the name Sphaerisporangium siamense sp. nov. is proposed.
Description of Sphaerisporangium siamense sp. nov.
Sphaerisporangium siamense: si.a.men'se. NL neut adj. siamense, of or belonging to Siam, the old name of Thailand, the source of the soil from which the type strain was isolated.
Gram-positive, non-acid–alcohol-fast, non-motile actinomycete. Pale pink to pale orange substrate mycelium is produced on ISP medium 2, oatmeal-nitrate agar and yeast-starch agar with white aerial mycelia. Light-yellowish soluble pigments are produced in ISP medium 3 and oatmeal-nitrate agar. Spherical spore vesicles are borne on aerial mycelia. The pH range for growth is pH 5.0–9.0. Cells can grow in the presence of 0–2% NaCl. Catalase, oxidase and H2S production is positive. Allantoinase and nitrate reduction is negative. Utilization of raffinose, D(+)cellobiose, L(−)fucose, D(+)galactose, D(+)glucose, lactose, maltose, D(+)mannose and D(−)ribose is positive. Utilization of adonitol, arabinose, D(−)fructose, myo-inositol, D(−)mannitol, melibiose, L(−)rhamnose, D(−)sorbitol, sucrose, xylitol and D(+)xylose is negative. Casein, hypoxanthine, L-tyrosine and starch are degraded. Aesculin, allantoin, arbutin, cellulose, guanine, xanthine and xylan are not degraded. Acid phosphatase, alkaline phosphatase, aminopeptidase, esterase C4, α-glucosidase, β-glucosidase, leucine, lipase C8, phosphoamidase, trypsin and valine aminopeptidase are detected using the API ZYM enzyme assay; chymotrypsin, cystine aminopeptidase, α-fucosidase, α-galactosidase, β-galactosidase, β-glucuronidase, lipase C14, α-mannosidase and N-acetyl-β-glucosaminidase are negative. The diagnostic diamino acid of the peptidoglycan is meso-diaminopimelic acid. Whole-cell sugars are galactose, glucose, madurose, mannose and ribose. The glycan moiety of the murein is acetylated. Major phospholipids are diphosphatidylglycerol, phosphatidylethanolamine and phosphatidylinositol mannoside. The predominant menaquinones are MK-9, MK-9(H2) and MK-9(H4). Mycolic acids are not detected. The major fatty acid in cellular fatty acids profile is iso-C16: 0. The G+C content of the type strain DNA is 71.9 mol%.
The type strain, strain SR14.14T (=BCC 41491T=NRRL B-24805T=NBRC 107570T), was isolated from the rhizospheric soil of rubber tree, collected from Phatthalung province, Thailand.
Accession codes
References
Ara, I. & Kudo, T. Sphaerosporangium gen. nov., a new member of the family Streptosporangiaceae, with descriptions of three new species as Sphaerosporangium melleum sp. nov., Sphaerosporangium rubeum sp. nov. and Sphaerosporangium cinnabarinum sp. nov., and transfer of Streptosporangium viridialbum Nonomura and Ohara 1960 to Sphaerosporangium viridialbum comb. nov. Actinomycetologica 21, 11–21 (2007).
Cao, Y.- R., Jiang, Y, Xu, L.- H. & Jiang, C.- L. Sphaerisporangium flaviroseum sp. nov. and Sphaerisporangium album sp. nov., isolated from forest soil in China. Int. J. Syst. Evol. Microbiol. 59, 1679–1684 (2009).
Stackebrandt, E., Rainey, F. A. & Ward-Rainey, N. L. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int. J. Syst. Bacteriol. 47, 479–491 (1997).
Zhi, X.- Y., Li, W.- J. & Stackebrandt, E. An update of the structure and 16S rRNA gene sequence-based definition of higher ranks of the class Actinobacteria, with the proposal of two new suborders and four new families and emended descriptions of the existing higher taxa. Int. J. Syst. Evol. Microbiol. 59, 589–608 (2009).
Nonomura, H. & Ohara, Y. Distribution of the actinomycetes in soil. V. The isolation and classification of the genus Streptosporangium. J. Ferment. Technol. 38, 405–409 (1960).
Küster, E. & Williams, S. T. Media for the isolation of streptomycetes: starch casein medium. Nature 202, 928–929 (1964).
Shirling, E. B. & Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340 (1966).
Jacobson, E., Grauville, W. C. & Fogs, C. E. Color Harmony Manual 4th edn (Container Corporation of America, Chicago, IL, 1958).
Gordon, R. E., Barnett, D. A., Handerhan, J. E. & Pang., C. H.- N. Nocardia coeliaca, Nocardia autotrophica, and the nocardin strain. Int. J. Syst. Bacteriol. 24, 54–63 (1974).
Gordon, R. E. & Mihm, J. M. A comparative study of some strains received as nocardiae. J. Bacteriol. 73, 15–27 (1957).
Becker, B., Lechevalier, M. P. & Lechevalier, H. A. Chemical composition of cell-wall preparations from strains of various form-genera of aerobic actinomycetes. Appl. Microbiol. 13, 236–243 (1965).
Hasegawa, T., Takizawa, M. & Tanida, S. A rapid analysis for chemical grouping of aerobic actinomycetes. J. Gen. Appl. Microbiol. 29, 319–322 (1983).
Uchida, K. & Aida, K. An improved method for the glycolate test for simple identification of the acyl type of bacterial cell walls. J. Gen. Appl. Microbiol. 30, 131–134 (1984).
Minnikin, D. E., Patel, P. V., Alshamaony, L. & Goodfellow, M. Polar lipid composition in the classification of Nocardia and related bacteria. Int. J. Syst. Bacteriol. 27, 104–117 (1977).
Collins, M. D., Pirouz, T., Goodfellow, M. & Minnikin, D. E. Distribution of menaquinones in actinomycetes and corynebacteria. J. Gen. Microbiol. 100, 221–230 (1977).
Tomiyasu, I. Mycolic acid composition and thermally adaptative changes in Nocardia asteroides. J. Bacteriol. 151, 828–837 (1982).
Tamaoka, J. & Komagata, K. Determination of DNA base composition by reversed-phase high-performance liquid chromatography. FEMS Microbiol. Lett. 25, 125–128 (1984).
Ezaki, T., Hashimoto, Y. & Yabuuchi, E. Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int. J. Syst. Bacteriol. 39, 224–229 (1989).
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F. & Hopwood, D. A. Practical Streptomyces Genetics (John Innes Foundation, Norwich, 2000).
Duangmal, K., Ward, A. C. & Goodfellow, M. Selective isolation of members of the Streptomyces violaceoruber clade from soil. FEMS Microbiol. Lett. 245, 321–327 (2005).
Lane, D. J. in 16S/23S rRNA Sequencing (eds Stackebrandt, E. & Goodfellow, M.) 115–148 (John Wiley & Sons, Chichester, 1991).
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids. Res. 25, 4876–4882 (1997).
Fitch, W. M. & Margoliash, E. Construction of phylogenetic trees: a method based on mutation distances as estimated from cytochrome c sequences is of general applicability. Science 155, 279–284 (1967).
Fitch, W. M. Towards defining the course of evolution: minimum change for a specific tree topology. Syst. Zool. 20, 406–416 (1971).
Saitou, N. & Nei, M. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Felsenstein, J. PHYLIP (Phylogenetic Inference Package) Version 3.5c. Distributed by the author. (University of Washington, Seattle, WA, 1993).
Van de Peer, Y. & De Wachter, R. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows Environment. Comp. Appl. Biosci. 10, 569–570 (1994).
Page, R. D. M. TreeView: an application to display phylogenetic trees on personal computers. Comp. Appl. Biosci. 12, 357–358 (1996).
Wayne, L. G. et al. International Committee on Systematic Bacteriology. Report of the ad hoc committee on the reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37, 463–464 (1987).
Acknowledgements
We thank Professor JP Euzéby for his kind advice on naming the species. This study was supported by the Thailand Research Fund, Commission on Higher Education and Kasetsart University Research and Development Institute.
Author information
Authors and Affiliations
Corresponding author
Additional information
Accession code
The GenBank accession number for the 16S rRNA gene sequence of strain SR14.14T is HM043727.
Rights and permissions
About this article
Cite this article
Duangmal, K., Mingma, R., Pathom-aree, W. et al. Sphaerisporangium siamense sp. nov., an actinomycete isolated from rubber-tree rhizospheric soil. J Antibiot 64, 293–296 (2011). https://doi.org/10.1038/ja.2011.17
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2011.17
Keywords
This article is cited by
-
Sphaerisporangium fuscum sp. nov., Isolated from Sediment of Anmucuo Lake in Tibet Autonomous Region of China
Current Microbiology (2022)