Abstract
The variability in genome content among closely related strains of prokaryotes has been one of the most remarkable discoveries of genomics. One way to approach the description of this so-called pan-genome is to compare one reference strain genome with metagenomic sequences from the environment. We have applied this approach to one extreme aquatic habitat, saturated brines in a solar saltern. The genome of Haloquadratum walsbyi strain DSM 16790 was compared to an environmental metagenome obtained from the exact site of its isolation. This approach revealed that some regions of the strain genome were scarcely represented in the metagenome. Here we have analyzed these genomic islands (GI) in the genome of DSM 16790 and compared them with the complete sequence of some fosmids from the environmental library. Two of the islands, GI 2 and GI 4, overlapped with two large guanine and cytosine (GC)-rich regions that showed evidence of high variability through mobile elements. GI 3 seemed to be a phage or phage-remnant acquired by the reference genome, but not present in most environmental lineages. Most differential gene content was related to small molecule transport and detection, probably reflecting adaptation to different pools of organic nutrients. GI 1 did not possess traces of mobile elements and had normal GC content. This island contained the main cluster of cell envelope glycoproteins and the variability found was different from the other GIs. Rather than containing different genes it consisted of homologs with low similarity. This variation might reflect a phage evasion strategy.
Similar content being viewed by others
Introduction
One of the major challenges for understanding prokaryotic diversity and evolution is the notable heterogeneity of genomes that can be found within a single species or operational taxonomic unit. Studies carried out mostly with pathogenic isolates (and sometimes non-pathogenic relatives) have revealed highly dynamic genomes indeed (Tettelin et al., 2005; Dempsey et al., 2006; Hochhut et al., 2006; Petrosino et al., 2006; Willenbrock et al., 2006). Bacterial genomes can change dramatically in size, gene repertoire and synteny among the different strains or environmental lineages characterized as belonging to specific, well-defined taxa (Lerat et al., 2005; Thompson et al., 2005; Dempsey et al., 2006; Green and Bohannan, 2006). The term pan-genome was coined to describe the gene repertoire carried by a well-defined species (Tettelin et al., 2005). Most of the information regarding the pan-genomic structure of prokaryotic species derives from the comparative genomics of multiple isolates from a single species (Tettelin et al., 2005; Read and Ussery, 2006). However, this approach has limitations: the sequenced strains may not represent the actual diversity of the species. For example, clinical isolates are representatives of highly virulent lineages selected by the defense systems of the host or by antibiotic resistance. Free-living cells are selected during cultivation by their ability to grow in the artificial environment of the laboratory and may not accurately depict the environmental population. An alternative approach for studying the pan-genome is to use metagenomic sequence data obtained directly from an environment in which a species is known to be well represented. The metagenome contains sequences from the different individuals of the species and this information can be used to infer genome diversity among these lineages. At least one reference strain genome has to be available to identify the metagenomic fragments as belonging to the species under study.
In a recent paper (Coleman et al., 2006), this approach was used to study genomic diversity in Prochlorococcus marinus, a marine cyanobacterium predominant in open ocean oligotrophic waters. By comparing a strain genome with shotgun metagenomic data from the Sargasso Sea, they identified well-defined regions of the reference genome that had very few or no homologous sequences in the metagenome. These regions, called Genomic Islands (GI), are considered hypervariable and indeed may be unique to the reference strain, as they contain telltale features that indicated phage-mediated lateral gene transfer. Many GI genes were related to nutrient stress and different light intensity adaptation. We have applied a similar approach to an extremely simplified aquatic habitat, saturated brines in a solar saltern. This is one of the most challenging extreme environments and the communities that thrive there are typically dominated by an assemblage of two prokaryotic species: the hyperhalophilic bacterium Salinibacter ruber and its archaeal counterpart, notorious for its peculiar postal stamp morphology, Haloquadratum walsbyi. The genome of H. walsbyi DSM 16790 was described recently (Bolhuis et al., 2006). In a previous work (Legault et al., 2006), an environmental saltern fosmid library was constructed from DNA extracted from cells enriched with the H. walsbyi morphology. End-sequence analysis of the fosmid inserts revealed a remarkable diversity of genes, evidence for GIs, and established that H. walsbyi species gene pool was, even in that relatively simple and constant environment, at least twice the genome size of the sequenced strain DSM 16790. Although the case was made for certain gene types to be found in the adaptive pool, the relatively limited data provided by fosmid end sequencing precluded an in-depth analysis. Here, we identified the GIs present in the H. walsbyi DSM 16790 genome and compared them with those of the complete sequences of environmental fosmids. The differences found have implications for the potential eco-physiology of H. walsbyi. A picture of extreme diversity and plasticity arises that contrasts with other simplified extreme environments analyzed before by similar approaches (Tyson et al., 2004).
Materials and methods
Genomic library and sequencing of fosmids clones
The environmental genomic library used here was constructed as described in Legault et al. (2006). Twenty-three fosmids were chosen and sequenced accordingly with the details described in Results. The length varies between 10 480 and 38 210 pb (Figure S1). Clone 2B07 was fully sequenced (Genome Express, Meylan, France) using a MegaBase 4000 capillary sequencer (Amersham Biosciences, Piscataway, NJ, USA). The remaining eHwalsbyi fosmids were sequenced by pyrosequencing 454 technology (454 Life Sciences, Branford, CT, USA). A total of 540 945 pb were assembled.
Annotation of fosmids
Protein coding genes were predicted using the annotation package GLIMMER (Delcher et al., 1999), and were further manually curated. Spacers were subsequently searched against the non-redundant database using basic local alignment search tool (BLAST) (Altschul et al., 1990) to ensure that no open reading frame (ORF) had been missed. ORFs were compared to known proteins in the non-redundant database (http://www.ncbi.nlm.nih.gov/) using the BLASTX program (translated DNA vs protein). All hits with an e-value greater than 10−5 were considered non-significant. For each sequence where a significant hit could be found, another round of BLAST was performed on parts of the sequence not covered by the best BLAST hit. Preliminary analysis of these additional gene fragments showed that they did not impact the results significantly and were not considered in the final analyses. GC content was identified using the ‘geecee’ program from EMBOSS package (Rice et al., 2000). DNA similarity comparisons were performed using the BLASTN program between the fosmids and the fosmid-ends with DSM 16790.
Sequence analysis
Alignments were generated using MUSCLE version 3.6 (Edgar, 2004) and ClustalW (Thompson et al., 1994) and edited manually as necessary (Chenna et al., 2003). Phylogenetic analysis of proteins was performed using the MEGA3 phylogenetic tool software package (Kumar et al., 2004). MUMmer analysis: 2947 sequences from saltern crystallizer metagenome (accession numbers DU826964–DU824018) were aligned against reference genome of Haloquadratum walsbyi DSM 16790 using the MUMmer program version 3.19 (Kurtz et al., 2004). Specifically, we used the ‘promer’ program with the maxmatch option to calculate alignments and the ‘mummerplot’ program to generate the percent identity plot depicted in Figure 1. ACT analysis: the Artemis Comparison Tool (ACT, Release 5, The Sanger Institute (Carver et al., 2005)) allowed an interactive visualization of comparisons between the complete genome and metagenome-related sequences. The comparison data was generated by performing a BLASTN search of the metagenome sequences against the DSM 16790 genome, which allowed identifying regions of similarity, insertions and rearrangements that were comparable to previously characterized genome DSM 16790.
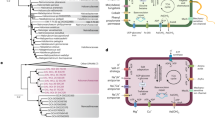
Haloquadratum walsbyi DSM 16790 genome and Genomic Islands. (a) GC-content of H. walsbyi genome plotted with a sliding window of 1000 nucleotides. The position of hmu* (halomucin), an unusually high %GC gene, is shown with an arrow. Location of integrases and IS transposases along the genome are indicated. (b) Fosmid-end coverage. Individual fosmid-end sequences were aligned to the sequenced strain genome and the alignment-sequence conservation visualized in the form of percent identity plot. Each dot on the graph represent an individual fosmid-end sequence aligned along its homologous region in H. walsbyi DSM 16790 genome. Y axis reflects its nucleotide percent identity to the syntenic region. The regions with unusually low representation in the metagenome are shaded and described in the text as genomic islands (GI). (c) Location of environmental H. walsbyi (eHwalsbyi) fosmids that have been fully (or partially) sequenced. We define as syntenic a region of a fosmid that contains more than 50% of its genes in the same relative location and orientation as in the reference genome. Syntenic fosmids are represented in green. The non-syntenic fosmids are represented in red and are connected to the genome with lines of variable thickness. The thickness of lines is proportional to the size of the segments with significant similarity to the DSM 16790 genome.
Accession numbers
The sequences have been submitted to GenBank under the accession numbers DQ314492 and EF583981–EF584002. The sequence of the complete genome of Haloquadratum walsbyi DSM 16790 is available under GenBank accession numbers AM180088.1 (chromosome) and AM180089.1 (plasmid PL47).
Results
The similarity of the fosmid-end sequences of the metagenomic library to the H. walsbyi DSM 16790 genome is depicted in Figure 1b. The comparison revealed that nucleotide sequences identical to most of the reference genome were present in the environment. However, certain regions of the DSM 16790 genome were less represented in the metagenome (which we defined as GIs). These have been interpreted previously as regions specific to certain lineages or strains (Coleman et al., 2006). Figure S3 presents a more detailed depiction of the similarity and representation of GI 1 among the fosmid-end sequences.
Twenty-three metagenomic fosmids belonging to environmental H. walsbyi or its close relatives (following the nomenclature introduced in Coleman et al. we refer to these sequences as eHwalsbyi) were fully (or nearly fully) sequenced to better understand their genomic diversity in the environment. We selected eHwalsbyi fosmids for sequencing on the grounds that end sequences indicated discontinuity with the strain genome: (i) 12 fosmid inserts had both ends exhibiting homology to the DSM 16790 genome at distances much larger than the fosmid insert size, (ii) 6 fosmid inserts had just one end exhibiting high similarity to the DSM 16790 genome, and (iii) 5 fosmid inserts had no sequence similarity to the reference genome, but both ends displayed low GC-content (low %GC is indicative of H. walsbyi (Bolhuis et al., 2006; Legault et al., 2006)).
The distribution of the selected fosmids along the DSM 16790 genome was determined through direct sequence comparison by BLASTN and alignment of the sequences using Artemis Comparison Tool. Twenty-one fosmids were positioned by this method and a graphic view of their location is shown in Figure 1c. Two fosmids (eHwalsbyi 011 and 022, Supplementary Figure S1) did not demonstrate any similarity to the DSM 16790 genome. Four fosmids (eHwalsbyi 058, 101, 416 and 420) appeared completely syntenic to their corresponding sections of the strain genome. The combined total sequence from these fosmid inserts (65 kpb) had an average nucleotide identity of 99.3% and, in particular, fosmid eHwalsbyi 416 had 100% identity over 90% of the 24 886 sequenced nucleotides. Together, they illustrate that H. walsbyi lineages contain some regions of highly conserved gene order and sequence. Actually, even fosmids that were not syntenic had an average similarity of ca. 98% to the homologous parts of the DSM 16790 genome. Of the sequenced fosmids, only three appeared clearly associated with GIs (eHwalsbyi 559, 485, 033), but many of the remaining 16 fosmids were located close to the GIs or contained genes similar to those found in GIs 2 and 4. With the exceptions noted above, all eHwalsbyi fosmids had some non-syntenic parts, that is, genes that were not present in the corresponding stretches of the DSM 16790 genome or appeared at different positions (Figure S1). In the following sections, we will focus on the fosmids found at or near the GIs. For the complete description of the eHwalsbyi fosmids and the genes in the non-syntenic regions, see Supplementary Information Table S1 and Figures S1 and S2.
GI 1 and fosmid eHwalsbyi 559
GI 1 (Figure 2) is located between nucleotides 257397–302834 (45.4 kpb) of DSM 16790 genome. This island is atypical compared to other GIs for two important reasons: first, the average GC-content (47.86%) is similar to the average of the genome (47.90%) and second, GI 1 is devoided of IS elements and putative phage genes (the transposase HQ1109 inside GI 1 is not functional). The main feature of GI 1 is a large cluster of cell-surface glycoprotein genes (CSG) (Schaffer and Messner, 2001; Schaffer et al., 2001), many of which could be components of the S layer that provides rigidity to the cell envelope of haloarchaea (Mengele and Sumper, 1992; Schaffer and Messner, 2001). GI 1 contains half of the 14 annotated CSGs in the DSM 16790 genome and a probable cell-surface adhesin. Bioinformatic analysis of CSGs present in GI 1 indicates that the ORF HQ1207, is most similar to the major S-layer component in Halobacterium salinarum and Haloarcula marismortui ATCC 43049 and may play a similar role in H. walsbyi (Blaurock et al., 1976; Trachtenberg et al., 2000). The relatively large number of CSGs found in H. walsbyi reference genome seems atypical since all sequenced haloarchaeal genomes have two or less (Bolhuis et al., 2006). This has been attributed to the complex architecture associated with the square cell morphology (Walsby, 2005). An alternative explanation is related to the demography of this organism. H. walsbyi cells reach extremely high population densities in the mature crystallizer community reaching up to 108 cells/ml. This high population density makes an ideal target for phage predation (Guixa-Boixereu et al., 1999), and the CSGs might act as recognition and/or attachment targets for phages.
GI 1 and fosmid eHwalsbyi 559. Yellow shadings link homologous regions between the island and the fosmid, and percent identity is shown in bold numbers. Location of G1 on H. walsbyi DSM 16790 genome is indicated by nucleotide position numbers at the beginning and the end. ORF names are as annotated in GenBank and are designated near each box.
The above hypothesis is supported by the sequence found in fosmid eHwalsbyi 559 (36 124 pb) that overlaps with most of GI 1 and contains an alternative cluster of CSGs (Figure 2 and Table S1). Although the fosmid had high synteny and overall sequence similarity to the DSM 16790 genome, there were two regions of variation. One is hmu2 gene (HQ1197) that codes for a CSG with similarity to the halomucin hmu gene (HQ1081) (see Figure 1a), and could be part of the cell envelope outer layer or capsule (Bolhuis et al., 2006); the other is ORF HQ1205, annotated as cell-surface adhesin that had the hallmarks of a secreted glycoprotein. Identity between the strain copies of hmu2 and HQ1205 and their environmental counterparts is only 24% at the nucleotide level (Figure 2). Similarly, located in GI 4 (see below) and nearly symmetric with respect to the replication origin are paralogous copies of hmu2 and two adjacent genes. Recombination involving these two paralogous clusters could explain the extremely high level of variation found in these genes.
GI 2 and fosmid eHwalsbyi 485
GI 2 (Figure 3) is located on the DSM 16790 chromosome between nucleotides 1272280–1457646 (185.3 kpb) and has several typical features of a hypervariable GI. One conspicuous indication is that it contained a high %GC region (1351161–1409095) with subregions as high as 57%, a value typical to phages and insertion sequence (IS) elements of H. walsbyi and other hyperhalophiles. The island is also rich in mobile elements: there are three functional IS-1341 (the predominant IS element in this genome) transposases, a probable integrase gene (PhiCh1 Int1-like), several helicase genes that appear to be phage-related (three rad/rad25 genes and one uvrD gene) and a probable helicase family protein. Other notable features of this GI were a low average coding-region density (65.2% compared with 74.5% for the whole DSM 16790 genome) and 14 pseudogenes. All these are hallmarks of a highly unstable genomic region.
GI 2 and fosmid eHwalsbyi 485. For notations see the legend for Figure 2.
Interestingly, this region also contains many genes involved in the transport of nutrients across the membrane and likely confers ecological adaptation or specialization. This island contains one of the six livHMGF(J) operons found in the DSM 16790 genome. Another cluster is found in GI 3 (see below). These genes are described as transporters of branched amino acids (leucine, valine and isoleucine). They are widely distributed in prokaryotes and it has been suggested that this transporter family's success lies in the diversification of their substrate specificities, capturing also other hydrophobic molecules such as lignin monomers, fatty acids and dicarboxylic acids derived from oils and fats (Larimer et al., 2004). Another gene cluster found in the island is the TRAP type C4-dicarboxylic acid permease cluster, also implicated in the transport of organic nutrients. Finally, a cluster involved in nitrate/nitrite transport (narK) and dissimilative nitrate reduction (respiration) to ammonia (narB and nirA) was also found within the island.
Fosmid eHwalsbyi 485 (31 408 pb) had some synteny to the beginning of GI 2 and illustrates the high variability that affects this genomic region. Of its 25 ORFs, 10 had extremely high nucleotide identity to those found in DSM 16790 (average, 98%; range, 95–100%) and they occurred in the same order and orientation (Figure 3, Table S1). On the other hand, there are notable differences. On eHwalsbyi 485, both copies of IS1341, two histidine kinase genes (HQ2053 and HQ2054), and a gene cluster probably involved in quorum sensing (homoserine lactone efflux) were absent. These genes were replaced on eHwalsbyi 485 by sn-glycerol-3-phosphate transport system (cluster ugp CEAB) and a different histidine kinase gene. Glycerol, the compatible solute produced by the green alga Dunaliella sp. (Phadwal and Singh, 2003; He et al., 2007) is probably one of the most abundant nutrients in this habitat. These glycerol transport genes have higher similarity to homologs found in H. marismortui and Halobacterium NRC1 than to a cluster of glycerol transport genes found elsewhere in the DSM 16790 genome (HQ1989–1992).
GI 3 and liv gene clusters
Located at 2602766–2661746 (58.9 kpb), GI 3 had two subregions (Figure 4). The first subregion (ca. 40 kpb), occupying a majority of the GI, is characterized by high proportion of hypothetical proteins, phage integrases, cdc6 homologs and bacterial conjugation-related genes within a high %GC region. This subsection of the island may be the remnant of a lysogenic phage inserted in the DSM 16790 genome and absent in many or most cells in the natural environment, as there were no identifiable homologs in the metagenome. The second subregion (ca. 20 kpb) comprises of another liv gene cluster (see above), a series of IS-1341 transposases and a return to average DSM 16790 genome %GC values.
GI 3. For notations see the legend for Figure 2.
From the data, it appears that livJ in H. walsbyi has a rich evolutionary history. The eHwalsbyi 464 liv cluster was most similar (over 97% nucleotide identity) to those found starting at position 2262733 in the DSM 16790 genome with the exception of the binding substrate subunit LivJ. An alignment of the HQ2969 versus the eHwalsbyi 464 LivJ subunits showed three rearranged fragments with nearly identical sequence (Figure 5a): the HQ2969 N-terminus, which is a putative membrane lipoprotein lipid attachment site, and the central section were flipped with the C-terminal part in the environmental version. Interestingly, the eHwalsbyi 464 livJ was more similar to the paralog located in GI 2 (41%, see above) but had the same fragment order as the LivJ in GI 3. The variation in domain order may have resulted in a non-functioning protein or perhaps in a change in substrate binding, although the chemical nature of the substrate might remain similar. There is an association between the evolutionary history of livJ and IS element IS1341. In Figure 5b, a phylogenetic tree of all H. walsbyi LivJ proteins showed that HQ2192 (GI 2), HQ2809, HQ3303 (GI 3), HQ2754 and eHwalsbyi 464 livJ form a cluster. Near or adjacent to these genes were copies of the IS element IS1341 with the only exception of HQ2809; however, there was putative evidence of an IS element, as there exists a small ORF similar to the zinc-binding domain characteristic of this transposase. These data suggest that IS 1341 is involved in the spread and variation of the livJ gene, which might provide a diversity of transport specificities and therefore ecological adaptations for the organisms that carry them.
livHMGF(J) containing fosmid eHwalsbyi 464 and LivJ phylogeny. (a) Fosmid eHwalsbyi 464 and syntenic region in H. walsbyi DSM 16790 genome. For notations, see the legend for Figure 2. (b) Phylogenetic tree of all the genome and metagenome substrate-binding LivJ proteins.
GI 4 and fosmid eHwalsbyi 033
GI 4 (Figure 6, Table S1), located between 2799525 and 3012525 (213 kpb), is extremely rich in transposases and putative phage-related genes and contains a large high %GC region. Within the first part of the island exists a cluster of glycoprotein genes (HQ3467, HQ3468 and HQ3469) that seem to be paralogous to HQ1197, HQ1196 and HQ1195 located in GI 1 (average nucleotide similarity of the genes present at both islands was ca. 40%). The first CSG in GI 4, annotated as halomucin 3 (hmu3), had no orthologous sequences among the sequenced fosmid ends, although it codes for a very large protein with 2079 amino-acid residues. Also, the hmu2 paralog found in GI 1 had only a couple of homologs of very low similarity in the metagenomic library (data not shown). The second and third glycoproteins of GI 4 had only one hit each at 26 and 99% similarity, respectively. Therefore, these genes appear remarkably variable among the members of H. walsbyi. It is possible that the variation affecting these putative CSG genes found in GI 1 and GI 4 is caused by intragenomic recombination, as both regions are equidistant from the origin of replication, a condition known to facilitate such phenomenon (Hughes, 2000; Mackiewicz et al., 2001).
GI 4 and fosmid eHwalsbyi 033. For notations, see the legend for Figure 2.
GI 4 contains many genes potentially involved in cell-envelope glycosylation and capsule biosynthesis, including a cluster involved in sialic-acid biosynthesis. Sialic acid is a common sugar found in the glycosidic moiety of mucins and is responsible for their extreme hydrophilicity (Schauer, 2000). It is interesting that in one of the P. marinus islands there is also a cluster of genes involved in sialic-acid synthesis (Schauer, 2000; Coleman et al., 2006). Finally, GI 4 also contains two adjacent clusters of ABC-type transporter genes involved in the translocation of small molecules, one involved in urea transport and other in cobalt transport, and one ABC-type transport system similar to a lipoprotein release factor.
None of the environmental fosmids sequenced was syntenic with GI 4. However, fosmid eHwalsbyi 033 (25 121 pb) contains rearranged GI 4 fragments (Figure 6). Additionally, this fosmid contained a few genes closely related to hypothetical proteins found in a plasmid of H. marismortui (pNG600). Another fosmid, eHwalsbyi 539 (32 713 pb), contains a large inversion with one end just at the beginning of GI 4 (three ORFs upstream) and might involve ca. 462 kpb (Figure S1 and Table S1).
Discussion
The saturated brines of solar salterns, highly enriched in magnesium salts, make life impossible for all but the most hyperhalophilic and specialized cells (Bolhuis et al., 2006). Molecular approaches revealed the mature crystallizer community to be largely dominated by H. walsbyi. Often, more than 80% of the dense biomass found in these waters is made of square archaea in which 16S rRNA genes differ by less than 1% (Bolhuis et al., 2004). Furthermore, in a previous work by sequencing only fosmid ends, we could find sequences highly similar to the DSM 16790 strain genome, about half of the sequences with the peculiar low GC content of H. walsbyi (Legault et al., 2006). However, the other half had low or no similarity to this genome. The short sequences at the fosmid ends did not allow us to establish whether the highly similar sequences found there were linked (that is, within the same lineage) to the dissimilar environmental sequences. In this work, we report that some regions of the DSM 16790 genome and the metagenome show a remarkable level of conservation, often close to 100% nucleotide identity. But the data presented here show that indeed the different lineages contain regions of high similarity interspaced with others of low or no similarity to the strain genome.
The first reports comparing metagenomic data with reference genomes were derived from the analysis of the acid-mine drainage biofilm (e.g., Tyson et al., 2004). There, genetic diversity was shown to be quite restricted even at the level of nucleotide substitutions among the different environmental genomes. As previously discussed (Legault et al., 2006), differences in our data with the acid-mine drainage biofilm should be expected. The chemolithotrophic prokaryotic assemblage of the acid-mine drainage fix CO2 and N2, which restrict the set of required resources. Contrastingly, the dominant organisms in the saturated brines are heterotrophs, which typically require a wide variety of carbon and other nutrients (during day time haloarchaea can derive energy from light by rhodopsins, but they cannot fix CO2 or N2). These nutrients are obtained from an extremely diverse set of organic compounds released by the massive Dunaliella sp. populations and other microbes that thrive at lower salinities (Pedros-Alio et al., 2000; Gasol et al., 2004). Thus, we suggest that different cells or lineages within H. walsbyi specialize in the exploitation of different organic compounds and coexist in such a chemically diverse set of resources. They do so by containing different gene pools that are largely associated with the GIs described here.
GI 2 and GI 4 have all the hallmarks of prokaryotic GIs (such as the pathogenicity islands that have been known for many years), that is, atypical GC-content and rich complement of mobile elements. However, the variability found in GI 1 was atypical in many ways. We did not find evidence of either phage or IS element involvement. Instead, intragenomic recombination could be the reason for the variability found there. If, as the evidence seems to indicate, GI 1 contains the genes required to synthesize the rigid components of the cell envelope, it is understandable that the region must be protected from excessive variation that could endanger the viability of the cell. Unfortunately, very little is known about the cell-envelope structure of H. walsbyi. Probably, the square shape requires a more diverse set of CSGs, which is reflected in the DSM 16790 genome. However, some of the CSG genes seem to be paralogous and might be involved in variability generation. Recombination among different functional copies provides variation with less risk of generating abortive cells. The recombination of lipopolysaccharide (LPS) cluster (rfb genes) in Salmonella enterica is a paradigm of this strategy (Xiang et al., 1994; Wang et al., 2002). In H. walsbyi, it probably reflects phage evasion by a ‘competitive dominant’ microdiversity (Thingstad, 2000). Electron microscopy studies in the Santa Pola crystallizer have shown that between 1% and 10% of the square cells are filled with lemon-shaped phage particles (Guixa-Boixereu, 1996). It is easy to imagine that the effects of such a dense phage population in this high-biomass low-diversity habitat could be catastrophic without an evasion strategy that promotes variations at the phage attachment sites (probably CSGs). The polysaccharide related genes found in GI 4 might contribute to a similar purpose (see below).
The most common variable feature found in H. walsbyi genomes is the complement of transporters, mostly of amino acids and peptides. About half of the biomass composition is protein and thus it seems appropriate that these transporters would be the main adaptations of the heterotrophic community that develops in saturated brines. It is reasonable to think that a single cell or clonal lineage could not utilize all the available biomass components and that different lineages within the same species should specialize themselves in the use of different compounds, particularly monomers, to coexist and avoid direct competition for resources. Another very common function assigned to genes within variable regions is the sensing part of two-component regulators. They might reflect again the specialization of different genomes in the use of different compounds that require alternative sensors for efficient regulation.
Remarkably, many of our findings are quite similar to those reported for P. marinus (Coleman et al., 2006). In both cases, metagenomic data were used to detect particularly labile regions in a prokaryotic genome. Although P. marinus and H. walsbyi are located at distant branches of the prokaryotic phylogenetic tree and are physiologically (phototroph versus heterotroph) and ecologically different (nutrient-limited, high-diversity versus extreme, low-diversity environment), both provide similar pan-genomic frameworks. We found some islands similar to P. marinus that clearly show evidence of phage remnants and are highly enriched in genes with no orthologs in the metagenome. Also similar were some of the gene functions found in the islands. We found variability at the level of glycoproteins and polysaccharides that are exposed in the cell envelope, and in P. marinus, there were variable components of the LPS, potential targets for phage attachment. We did not find an association of the islands with tRNA genes as was found in P. marinus and which are common in pathogenicity islands. This might be a bacterial (as opposed to archaeal) characteristic. On the other hand, IS elements were found at the ends of all islands and are highly enriched within them. An interesting parallel with P. marinus is that several copies of hli genes, which are essential to perform photosynthesis under a wide range of conditions, were found in the islands. In H. walsbyi, we found the liv gene clusters that might be responsible for amino-acid (or other substrate families) uptake and are likely to help the organism adapt to their heterotrophic lifestyle. The sequenced fosmids indicate that H. walsbyi genome varies unevenly, much more so than in the case of P. marinus. The syntenic regions found had an average nucleotide identity of 98%, which was higher than the 90% found for P. marinus. However, the degree of variability in non-syntenic regions and in the GIs is extremely high, and indicates that the gene pool from which H. walsbyi draws to fulfil its ecological requirements is not small. Even more remarkable is the fact that, as previously shown (Legault et al., 2006), most of this diverse gene pool has the genomic imprint of H. walsbyi, easy to distinguish from most haloarchaea or hyperhalophiles in general, owing to its low GC-content. In other words, this diverse gene pool is contained within the large but relatively homogeneous population (as defined by 16S rRNA gene divergence) of square archaea present in the crystallizer.
This work provides insight into prokaryotic species' genomic diversity in general. The small size of prokaryotic cells precludes them from having large genomes and consequently, reduces any individual from utilizing the spectrum of available resources. Individual cells are specialized to a specific set of nutritional requirements but do not directly compete with other members of the same species for the same resources. This is in striking contrast to the way species are typically conceived, where intraspecies competition is for the same resources. Instead, this evolutionary strategy is reminiscent of multicellular eukaryotes that have specialized cells performing different physiological functions within the same organism. However, in prokaryotes the entire gene repertoire is not included in a single cell but in independent lineages (or clonal descent lines) and its phages (Breitbart and Rohwer, 2005) and comprises the species pan-genome. The crystallizer metagenome also illustrates the central role played by phages in the biology of H. walsbyi and probably in most prokaryotes. Most non-syntenic fosmids had genes hinting of phage involvement. Of course, the nearly mono-specific community structure of saturated brines provides a nearly ideal setting for carrying large phage populations. However, recent studies of marine prokaryotes that live in comparatively diluted environments provide similar interpretations (Sullivan et al., 2005; Angly et al., 2006).
Accession codes
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ . (1990). Basic local alignment search tool. J Mol Biol 215: 403–410.
Angly FE, Felts B, Breitbart M, Salamon P, Edwards RA, Carlson C et al. (2006). The marine viromes of four oceanic regions. PLoS Biol 4: e368.
Blaurock AE, Stoeckenius W, Oesterhelt D, Scherfhof GL . (1976). Structure of the cell envelope of Halobacterium halobium. J Cell Biol 71: 1–22.
Bolhuis HH, Palm PP, Wende AA, Falb MM, Rampp MM, Rodriguez-Valera FF et al. (2006). The genome of the square archaeon Haloquadratum walsbyi: life at the limits of water activity. BMC Genomics 7: 169.
Bolhuis H, Poele EM, Rodriguez-Valera F . (2004). Isolation and cultivation of Walsby's square archaeon. Environ Microbiol 6: 1287–1291.
Breitbart M, Rohwer F . (2005). Here a virus, there a virus, everywhere the same virus? Trends Microbiol 13: 278–284.
Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J . (2005). ACT: the artemis comparison tool. Bioinformatics 21: 3422–3423.
Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG et al. (2003). Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31: 3497–3500.
Coleman ML, Sullivan MB, Martiny AC, Steglich C, Barry K, Delong EF et al. (2006). Genomic islands and the ecology and evolution of prochlorococcus. Science 311: 1768–1770.
Delcher AL, Harmon D, Kasif S, White O, Salzberg SL . (1999). Improved microbial gene identification with GLIMMER. Nucleic Acids Res 27: 4636–4641.
Dempsey MP, Nietfeldt J, Ravel J, Hinrichs S, Crawford R, Benson AK . (2006). Paired-end sequence mapping detects extensive genomic rearrangement and translocation during divergence of Francisella tularensis subsp. tularensis and Francisella tularensis subsp. holarctica populations. J Bacteriol 188: 5904–5914.
Edgar RC . (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113.
Gasol JM, Joint I, Kristine G, Gustavson K, Benlloch S, Díez B et al. (2004). Control of heterotrophic prokaryotic abundance and growth rate in hypersaline planktonic environments. Aquat Microb Ecol 34: 193–206.
Green J, Bohannan BJ . (2006). Spatial scaling of microbial biodiversity. Trends Ecol Evol 21: 501–507.
Guixa-Boixereu N . (1996). Viral lysis and bacterivory as prokaryotic loss factors along a salinity gradient. Aquat Microb Ecol 11: 213–227.
Guixa-Boixereu N, Lysnes K, Pedros-Alio C . (1999). Viral lysis and bacterivory during a phytoplankton bloom in a coastal water microcosm. Appl Environ Microbiol 65: 1949–1958.
He Q, Qiao D, Bai L, Zhang Q, Yang W, Li Q et al. (2007). Cloning and characterization of a plastidic glycerol 3-phosphate dehydrogenase cDNA from Dunaliella salina. J Plant Physiol 64: 214–220.
Hochhut B, Wilde C, Balling G, Middendorf B, Dobrindt U, Brzuszkiewicz E et al. (2006). Role of pathogenicity island-associated integrases in the genome plasticity of uropathogenic Escherichia coli strain 536. Mol Microbiol 61: 584–595.
Hughes D . (2000). Co-evolution of the tuf genes links gene conversion with the generation of chromosomal inversions. J Mol Biol 297: 355–364.
Kumar S, Tamura K, Nei M . (2004). MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5: 150–163.
Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C et al. (2004). Versatile and open software for comparing large genomes. Genome Biol 5: R12.
Larimer FW, Chain P, Hauser L, Lamerdin J, Malfatti S, Do L et al. (2004). Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat Biotechnol 22: 55–61.
Legault BA, Lopez-Lopez A, Alba-Casado JC, Doolittle WF, Bolhuis H, Rodriguez-Valera F et al. (2006). Environmental genomics of ‘Haloquadratum walsbyi’ in a saltern crystallizer indicates a large pool of accessory genes in an otherwise coherent species. BMC Genomics 7: 171.
Lerat E, Daubin V, Ochman H, Moran NA . (2005). Evolutionary origins of genomic repertoires in bacteria. PLoS Biol 3: e130.
Mackiewicz P, Mackiewicz D, Kowalczuk M, Cebrat S . (2001). Flip-flop around the origin and terminus of replication in prokaryotic genomes. Genome Biol 2: interactions 1004.1–1004.4.
Mengele R, Sumper M . (1992). Drastic differences in glycosylation of related S-layer glycoproteins from moderate and extreme halophiles. J Biol Chem 267: 8182–8185.
Pedros-Alio C, Calderon-Paz JI, MacLean MH, Medina G, Marrase C, Gasol JM . (2000). The microbial food web along salinity gradients. FEMS Microbiol Ecol 32: 143–155.
Petrosino JF, Xiang Q, Karpathy SE, Jiang H, Yerrapragada S, Liu Y et al. (2006). Chromosome rearrangement and diversification of Francisella tularensis revealed by the type B (OSU18) genome sequence. J Bacteriol 188: 6977–6985.
Phadwal K, Singh PK . (2003). Effect of nutrient depletion on beta-carotene and glycerol accumulation in two strains of Dunaliella sp. Bioresour Technol 90: 55–58.
Read TD, Ussery DW . (2006). Opening the pan-genomics box. Curr Opin Microbiol 9: 496–498.
Rice P, Longden I, Bleasby A . (2000). EMBOSS: the European molecular biology open software suite. Trends Genet 16: 276–277.
Schaffer C, Messner P . (2001). Glycobiology of surface layer proteins. Biochimie 83: 591–599.
Schaffer C, Graninger M, Messner P . (2001). Prokaryotic glycosylation. Proteomics 1: 248–261.
Schauer R . (2000). Achievements and challenges of sialic acid research. Glycoconj J 17: 485–499.
Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW . (2005). Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol 3: e144.
Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL et al. (2005). Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial ‘pan-genome’. Proc Natl Acad Sci USA 102: 13950–13955.
Thingstad T . (2000). Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic ecosystems. Limnol Oceanogr 45: 1320–1328.
Thompson JD, Higgins DG, Gibson TJ . (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680.
Thompson JR, Pacocha S, Pharino C, Klepac-Ceraj V, Hunt DE, Benoit J et al. (2005). Genotypic diversity within a natural coastal bacterioplankton population. Science 307: 1311–1313.
Trachtenberg S, Pinnick B, Kessel M . (2000). The cell surface glycoprotein layer of the extreme halophile Halobacterium salinarum and its relation to Haloferax volcanii: cryo-electron tomography of freeze-substituted cells and projection studies of negatively stained envelopes. J Struct Biol 130: 10–26.
Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM et al. (2004). Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428: 37–43.
Walsby AE . (2005). Archaea with square cells. Trends Microbiol 13: 193–195.
Wang L, Andrianopoulos K, Liu D, Popoff MY, Reeves PR . (2002). Extensive variation in the O-antigen gene cluster within one Salmonella enterica serogroup reveals an unexpected complex history. J Bacteriol 184: 1669–1677.
Willenbrock H, Petersen A, Sekse C, Kiil K, Wasteson Y, Ussery DW . (2006). Design of a seven-genome Escherichia coli microarray for comparative genomic profiling. J Bacteriol 188: 7713–7721.
Xiang SH, Hobbs M, Reeves PR . (1994). Molecular analysis of the rfb gene cluster of a group D2 Salmonella enterica strain: evidence for its origin from an insertion sequence-mediated recombination event between group E and D1 strains. J Bacteriol 176: 4357–4365.
Acknowledgements
SCO acknowledges CAPES Foundation (Brazil) for the postdoctoral fellowship received. ABMC is supported by a MEC (Spain) Postdoctoral Fellowship. OZ is supported through a CIHR Postdoctoral Fellowship and is an honorary Killam Postdoctoral Fellow at Dalhousie University. This work was funded by the GEMINI (QLK3-CT-2002–02056) project of the European Commission, and MEC (Spain) (CTM2005–04564) ‘Metagenomic study of bacterioplankton of deep Mediterranean Sea’ project.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Rights and permissions
About this article
Cite this article
Cuadros-Orellana, S., Martin-Cuadrado, AB., Legault, B. et al. Genomic plasticity in prokaryotes: the case of the square haloarchaeon. ISME J 1, 235–245 (2007). https://doi.org/10.1038/ismej.2007.35
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2007.35
Keywords
This article is cited by
-
Distinct ecotypes within a natural haloarchaeal population enable adaptation to changing environmental conditions without causing population sweeps
The ISME Journal (2021)
-
Genomic variation and biogeography of Antarctic haloarchaea
Microbiome (2018)
-
Genomic variation in microbial populations inhabiting the marine subseafloor at deep-sea hydrothermal vents
Nature Communications (2017)
-
New insights into marine group III Euryarchaeota, from dark to light
The ISME Journal (2017)
-
Gene-specific selective sweeps in bacteria and archaea caused by negative frequency-dependent selection
BMC Biology (2015)