Abstract
Mesoaxial synostotic syndactyly with phalangeal reduction (MSSD) is a rare non-syndromic limb malformation with autosomal recessive inheritance. To date, only a few affected families with MSSD who had BHLHA9 mutations have been reported. The present report describes a consanguineous Pakistani family with five affected individuals with MSSD who exhibited an autosomal recessive pattern. Genotyping followed by Sanger sequencing was performed, and we identified a novel homozygous missense mutation (c.311T>C, p.Ile104Thr) in the BHLHA9 gene. This finding expands the spectrum of known mutations in the BHLHA9 gene that cause MSSD.
Similar content being viewed by others
Introduction
Development of the limbs during embryogenesis is a highly complex and well-organized process. During development, transcriptional regulation, numerous genes, and dosage-dependent signals are vigorously implicated in the regulation of molecular and cellular processes. The genes involved in early embryonic limb development include HOXD13, GLI3, GLI2, SHH, FGF8, WNT7A and WNT10B.1
Mesoaxial synostotic syndactyly with phalangeal reduction (MSSD, MIM 609432) is a rare congenital limb defect characterized by mesoaxial osseous synostosis at a metacarpal level, a reduction in the number of phalanges by one or more, hypoplasia of the distal phalanges of the preaxial and postaxial digits, clinodactyly of the fifth fingers, and preaxial fusion of the toes.2–4 Until now, only a couple of families with MSSD who exhibited autosomal recessive inheritance have been reported.2,4 The BHLHA9 gene (MIM 615416), encoding a basic helix-loop-helix (bHLH) A9 transcription factor and comprising a single exon, appears to be a master regulator gene for limb development because of its exclusive expression in the distal mesenchyme of the limb bud in the mouse embryo E10.5–E11.5.5–7 Studies have shown that BHLHA9 is associated with three different limb phenotypes: split-hand/foot malformation with long bone deficiency-3 (MIM 612576), complex camptosynpolydactyly (MIM 607539), and MSSD. So far, three missense mutations are known to be associated with MSSD, and there are three consanguineous family with MSSD who originated from Pakistan.4,8 Herein, we describe the fourth consanguineous family of Pakistani origin with MSSD. Sanger sequencing revealed a novel homozygous missense variant (c.311T>C, p.Ile104Thr) in the BHLHA9 gene that caused MSSD. This is the fourth MSSD-causing mutation in BHLHA9 and the first mutation in the helix domain.
Materials and methods
Ethical approval and family history
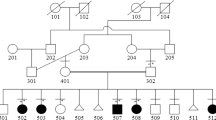
The study design was in accordance with the Helsinki Declaration and approved by the institutional review boards (IRB) of Peking Union Medical College (China), and Quaid-i-Azam University (Islamabad, Pakistan). The family resided in a suburb of the Khyber Pakhtunkhwa province, in Pakistan. The family had five individuals including three males (IV-1, IV-6 and IV-12) and two females (IV-3 and IV-8) who had MSSD (Figure 1a). In this family, there was no previous family history of limb malformations.
The pedigree structure and clinical characteristics of the proband. (a) The blackened symbols represent the affected individuals, and open symbols represent unaffected individuals. The arrow indicates the proband. The individuals who were available for genotyping are indicated by asterisks. (b) Photographs of the proband (IV-1) and affected individuals (IV-3, IV-8, and IV-12) show syndactyly phenotypes in the hands, as well as fusion of the second and third toes. (c) X-ray images of the proband’s right hand.
Genotyping and sequencing
Blood samples were obtained from five affected and five unaffected individuals in the family. Genomic DNA was extracted from peripheral blood samples using a QIAamp DNA Blood Midi Kit (Qiagen, Hilden, Germany) and quantified using a Nanodrop 2000 spectrophotometer (Thermo Scientific; Waltham, MA, USA). A single coding exon of the BHLHA9 gene (NM_001164405.1) was amplified using PCR. The PCR reaction was commenced with an initial 5-min denaturation step at 95 °C, followed by 35 cycles of denaturation (94 °C) for 30 s, annealing (56–62 °C) for 30 s, and extension (72 °C) for 30 s, and ended with a final extension step at 72 °C for 10 min. The PCR products were purified and subjected to direct sequencing using an ABI3700 autosequencer. Control samples from the 200 ethnically matched individuals were screened using Sanger sequencing.
Results
Clinical findings
As for proband IV-1, aged 24 years, four digits were observed on the right hand, and both hands had broadened thumbs. On the right hand, the third and fourth fingers were replaced by a single digit, which had a large proximal phalanx and a hypoplastic, club-shaped terminal phalanx, and the nails were reduced bilaterally. The fifth finger of the right hand exhibited clinodactyly. As for the left hand, the fourth finger had valgus deviations at the terminal phalanx. The phenotype of the feet was characterized by hallux varus; the partial appearance of zygodactyly between the second and third toes; and hypoplastic nails on the second, third, and fourth toes (Figure 1b). In addition to the previously described phenotype, the fifth toes of the proband’s younger sister (IV-3, aged 22 years) overlapped. As for his brother (IV-6, aged 16 years), his feet were involved, but his hands were normal. Subject IV-8 had five fingers on her right hand and four fingers on her left hand (Figure 1c). Upon a physical examination, subject IV-12 had four fingers bilaterally, but his feet were normal. An X-ray examination of the proband revealed synostosis of the third and fourth metacarpals, resulting in a large, single, and conical proximal phalanx, and dysplastic middle and terminal phalanges. The fifth fingers had clinodactyly with symphalangism of the middle and distal phalanges. The metacarpals had distal head hypoplasia (Figure 1c). There was no sign of bony involvement in the individual’s feet. A physical examination and roentgenographic studies of the other affected individuals in this family revealed a similar phenotype with a slightly different manifestation.
Mutation analysis
We identified a novel homozygous missense mutation at position 311 (c.311T>C) on the coding exon of BHLHA9 in all affected individuals using a Sanger sequencing analysis (Figure 2a). This missense transversion mutation led to the substitution of an isoleucine with a threonine at position 104 (p.Ile104Thr) that was predicted to be in the helix-loop-helix domain of a bHLH A9 transcription factor (Figure 2b). Genotyping in this family revealed that all affected individuals harbored the same homozygous c.311T>C missense mutation, whereas all obligatory carriers possessed the heterozygous mutation, suggesting a Mendelian co-segregation with the phenotype for all family members (Figure 1a). The novel mutation was not present in the dbSNP (http://www.ncbi.nih.gov/SNP) (accessed December 2015), 1000 genome project (the 1000 Genome Project Consortium 2015), or Exome Aggregation Consortium (ExAC) (http://www.exac.broadinstitute.org) and was not observed in 200 ethnically controlled individuals. The affected amino-acid residue is also been highly conserved throughout vertebrate evolution (Figure 2c). Use of different bioinformatics tools including Polyphen-2 (http://genetics.bwh.harvard.edu/pph2/), SIFT (http://sift.jcvi.org/), PROVEAN (http://www.provean.jcvi.org), and Mutation Taster (http://www.mutationtaster.org/) helped in the prediction that the mutation (c.311T>C) is pathogenic and affects protein structures.
The BHLHA9 mutation, protein structures, and conservation analysis of BHLHA9. (a) Genomic DNA sequencing electropherograms of the affected individual (upper), unaffected carrier (the middle), and control individual (lower). (b) Protein structures of BHLHA9. (*—amino-acid change reported by Malik et al.4 in MSSD and #—amino-acid change reported by Phadke et al.8 in Complex Camptosynpolydactyly), and the amino-acid change associated with the present family is shown using a red color in the diagram (**p.Ile104Thr). (c) The amino acids mutated in the affected individuals are conserved in different species. MSSD, mesoaxial synostotic syndactyly with phalangeal reduction.
Discussion
The bHLH proteins comprise a large superfamily of transcriptional regulators in organisms from yeast to humans and play an important role in developmental processes and sex determination, as well as in the development of the limbs, muscles, and nervous system. The bHLH family has two conserved domains, i.e., (1) a basic domain that binds to E-box and a consensus hexanucleotide sequence; and (2) the HLH domain that assists in the coordination of and interaction with the other protein for homodimer and heterodimer formation.9
The BHLHA9 gene locates on chromosome 17p13.3, and the encoded protein belongs to the bHLH superfamily and is involved in the limb developmental process. Klopocki et al. recognized that knockdown bhlha9 could cause pectoral fin shortening in zebrafish.5 In mouse models, Schatz et al.7 noticed the early expression of bhlha9 in the distal mesenchyme of limb buds of the E10.5-E11.5 mouse embryo, and homozygous bhlha9 knockout mice had various levels of syndactyly involving soft tissues, but not skeletal elements. Studies have shown that duplications or triplication on chromosome band 17p13.3 with a small region of overlap encompassing BHLHA9 is responsible for an autosomal dominant split-hand/foot limb malformation with long bone deficiency-3. Recently, Phadke et al. identified a novel pathogenic mutation in BHLHA9 that causes a complex camptosynpolydactyly characterized by grossly abnormal hands, polydactyly with digits arising from the dorsum of the hands, syn- and camptodactyly of some fingers, soft tissue syndactyly of the first and second toes, and hypoplastic nails.8,10 Malik et al.4 demonstrated that three neighboring missense mutations affecting the highly conserved DNA-binding region of BHLHA9 are associated with MSSD. Herein, we identified a novel homozygous missense variant (c.311T>C, p.Ile104Thr) in BHLHA9 in a consanguineous family of Pakistani origin. The mutation was not observed in 200 ethnically controlled individuals or in public databases (dbSNP142, 1000 Genomes, and ExAC), indicating that it is not a commonly silent polymorphism. The amino-acid residue Ile104 is highly conserved throughout vertebrate evolution, and four different bioinformatics tools predicted that the mutation (c.311T>C) is pathogenic and affects protein structures. This mutation is the first expected to affect the helix-loop-helix domain of BHLHA9. However, details regarding the pathogenic mechanism need to be further studied.
In summary, our observation, combined with those of other investigators, suggests that the BHLHA9 gene is a vital player in limb development. Additionally, our finding of the novel homozygous missense mutation (c.311T>C, p.Ile104Thr) expands our knowledge of the mutation spectrum of the BHLHA9 gene regarding limb phenotypes that helped in the prenatal genetic and molecular diagnosis of this family.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Moore KL, Persaud TVN, Torchia MG (eds). The developing human: Clinically oriented embryology. Elsevier Health Sciences: Philadelphia: USA, 2015.
Percin EF, Percin S, Egilmez H, Sezgin I, Ozbas F, Akarsu AN . Mesoaxial complete syndactyly and synostosis with hypoplastic thumbs: an unusual combination or homozygous expression of syndactyly type I. J Med Genet 1998; 35: 868–874.
Malik S, Arshad M, Amin-ud-Din M, Oeffner F, Dempfle A, Haque S et al. A novel type of autosomal recessive syndactyly: clinical and molecular studies in a family of Pakistani origin. Am J Med Genet A 2004; 126A: 61–67.
Malik S, Percin FE, Bornholdt D, Albrecht B, Percesepe A, Koch MC et al. Mutations affecting the BHLHA9 DNA-binding domain causes MSSD, mesoaxial synostoticsyndactyly with phalangeal reduction, Malik-Percin type. Am J Hum Genet 2014; 95: 649–659.
Klopocki E, Lohan S, Doelken SC, Stricker S, Ockeloen CW, Soares Thiele de Aguiar R et al. Duplication of BHLHA9 are associated with ectrodactyly and tibia hemimelia inherited in non-Mendelian fashion. J Med Genet 2012; 49: 119–125.
Nagata E, Kano H, Kato F, Yamaguchi R, Nakashima S, Takayama S et al. Japanese founder duplications/triplications involving BHLHA9 are associated with split-hand/foot malformation with or without long bone deficiency and Gollop-Wolfgang complex. Orphanet J Rare Dis 2014; 9: 125.
Schatz O, Langer E, Ben-Arie N . Gene dosage of the transcription factor Fingerin (bHLHA9) affects digit development and links syndactyly to ectrodactyly. Hum Mol Genet 2014; 23: 5394–5401.
Phadke SR, Kar A, Bhowmik AD, Dalal A . Complex camptosynpolydactyly and Mesoaxial synostotic syndactyly with phalangeal reduction are allelic disorders. Am J Med Genet A 2016; 170: 1622–1625.
Murre C, Bain G, van Dijk MA, Engel I, Furnari BA, Massari ME et al. Structure and function of helix loop helix proteins. Biochim Biophys Acta 1994; 1218: 129–135.
Phadke SR, Goutham P . Complex camptopolydactyly: An unusual hand malformation. Am J Med Genet 1999; 83: 191–192.
Acknowledgements
We would like to thank all individuals who participated in this study. This work was financially supported by the National Natural Science Foundation of China (NSFC) (Grant number: 81230015) and the Beijing Municipal Science and Technology Commission (Grant number: Z151100003915078).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Khan, A., Wang, R., Han, S. et al. A novel homozygous missense mutation in BHLHA9 causes mesoaxial synostotic syndactyly with phalangeal reduction in a Pakistani family. Hum Genome Var 4, 17054 (2017). https://doi.org/10.1038/hgv.2017.54
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/hgv.2017.54
This article is cited by
-
Genetic determinants of syndactyly: perspectives on pathogenesis and diagnosis
Orphanet Journal of Rare Diseases (2022)
-
Split hand/foot malformation with long bone deficiency associated with BHLHA9 gene duplication: a case report and review of literature
BMC Medical Genetics (2019)