Abstract
The R2 retrotransposon is here characterized in bisexual populations of the European crustacean Triops cancriformis. The isolated element matches well with the general aspects of the R2 family and it is highly differentiated from that of the congeneric North American Triops longicaudatus. The analysis of 5′ truncations indicates that R2 dynamics in T. cancriformis populations show a high turnover rate as observed in Drosophila simulans. For the first time in the literature, though, individuals harboring truncation variants, but lacking the complete element, are found. Present results suggest that transposition-mediated deletion mechanisms, possibly involving genomic turnover processes acting on rDNAs, can dramatically decrease the copy number or even delete R2 from the ribosomal locus. The presence of R2 does not seem to impact on the nucleotide variation of inserted 28S rDNA with respect to the uninserted genes. On the other hand, a low level of polymorphism characterizes rDNA units because new 28S variants continuously spread across the ribosomal array. Again, the interplay between transposition-mediated deletion and molecular drive may explain this pattern.
Similar content being viewed by others
Introduction
Non-LTR retrotransposons are transposable elements that can be either randomly distributed or inserted at a specific locus. Sequence specificity of insertion is considered an ancient strategy used by transposable elements to survive in the host genome by limiting their ability to disrupt essential genes (Malik et al., 1999). Ribosomal DNA represents a niche well exploited by non-LTR retrotransposons, and eight rDNA-specific families have been so far identified, with six of them inserting into the 28S gene (R1, R2, R4, R5, R6, RT; Eickbush and Eickbush, 2007).
One of the most studied non-LTR retrotransposon families is R2. Four clades have been so far recognized on the basis of the element's phylogeny and of the N-terminal zinc-finger motifs of the translated protein product (Kojima and Fujiwara, 2005). In particular, the R2-A, -C, and -D clades have 3, 2, and 1 zinc-finger motifs, respectively, whereas the N-terminal structure of the R2-B clade is, as yet, undetermined.
R2 occurs in the four triploblastic phyla Platyhelminthes, Arthropoda, Echinodermata, and Chordata, but its presence also in the diploblastic phylum Cnidaria suggests its vertical inheritance since the cladogenesis of Radiata and Bilateria (Kojima et al., 2006). Yet, R2 phylogeny is quite inconsistent with that of the host (Burke et al., 1999; Kojima and Fujiwara, 2005), only some taxonomic ‘subclades’ (sensu Kojima and Fujiwara, 2005; for example the Drosophila sp. and fishes–turtles subclades) being evident in the trees. Therefore, the hypothesis of horizontal transfer of the element has been put forward. However, its absence in taxa closely related to species harboring R2 indicates that the extinction of this retrotransposon has occurred several times, at least during insect and vertebrate evolution (for example, in Drosophila erecta, Drosophila orena, Fugu rubripes, mouse, and human). On the other hand, some species such as Popillia japonica and Ciona intestinalis have multiple lineages of the element (Burke et al., 1993; Eickbush et al., 1997; Kojima and Fujiwara, 2004). On the whole, both extinction and diversification can be explained by R2 evolutionary dynamics, showing a rapid turnover with high rates of retrotransposition and elimination (Pérez-Gonzalez and Eickbush, 2001, 2002; Zhang and Eickbush, 2005).
R2 inserts through a target primed reversed transcription mechanism (Christensen et al., 2006), which allows also the insertion of 5′-truncated copies; these are produced when the synthesis of R2 first-strand DNA is aborted before reaching the 5′ end of the element. The study of truncation variants is a tool to examine element activity: this aspect was deeply analyzed in laboratory stocks of Drosophila spp. In Drosophila simulans, in particular, a high turnover rate, together with transposition-mediated deletions, is responsible of the elimination of earlier generated truncation variants (Zhang et al., 2008). Moreover, the selective pressure against non-functional rDNA units tends to eliminate the R2-inserted copies through the unequal DNA exchanges acting in the concerted evolution of the ribosomal locus (reviewed in Nei and Rooney, 2005; Eickbush and Eickbush, 2007). Concerted evolution explains the variability pattern observed for repeated sequence families (such as ribosomal genes): the observed sequence variability within an evolutionary unit (a species, a subspecies or a population) is significantly lower than between different evolutionary units of the same rank. Concerted evolution is achieved through molecular drive, a process comprising the intragenomic homogenization of variants, through turnover mechanisms such as unequal crossing-over, gene conversion and rolling circle replication, and variant fixation within a group of reproductively linked bisexual organisms (Dover, 1982, 2002).
No detailed data are so far available about the effects of R2 insertion on sequence variability of 28S genes: in Drosophila, R2-inserted and non-inserted 28S units appear identical (Eickbush and Eickbush, 2007). However, in the crustacean Daphnia pulex, the DNA-mediated element Pokey determines a higher variability of the inserted 28S sequences with respect to those lacking the element (Penton and Crease, 2004; Glass et al., 2008).
We here characterize the R2 element in the Euroasiatic notostracan Triops cancriformis. The order Notostraca pertains to the class Branchiopoda, a primitive group of the Arthropoda sub-phylum Crustacea, recently placed within the Pancrustacea clade, strongly associated with Hexapoda (Halanych, 2004; Mallat et al., 2004). T. cancriformis is a well-known example of the very few living fossils: from its morphological stasis, it cannot be distinguished from the Triassic taxon T. cancriformis minor (Fisher, 1990). T. cancriformis inhabits ephemeral ponds and rice fields and shows a consistent variability in sexual reproductive strategies, which range from bisexuality (either gonochoric or hermaphroditic) to unisexuality (parthenogenesis; Mantovani et al., 2004, 2008). Here, we characterize the R2 element and analyze its turnover/elimination rates together with the 28S rDNA unit variation in Spanish gonochoric populations of T. cancriformis (sensu Korn et al., 2006).
Materials and methods
R2 molecular characterization and phylogenetic analysis
Tadpole shrimps were collected in Espolla (Spain): the same pond was sampled twice in 2004 and in 2006. Forty individuals were analyzed, 20 for each sampling (Table 1). Genomic DNA was extracted from single alcohol-preserved individuals with a standard phenol-chloroform protocol.
Samples were first checked for the presence of the R2 element using the forward degenerate primers described in Kojima and Fujiwara (2005), coupled with a 28SB-R reverse primer (Table 2), located 178 bp downstream of the element's insertion site. Of the tested primer pairs, only R2IF1>28SB-R gave an amplification product of the expected size (∼2000 bp); this was cloned and sequenced as described below. PCR amplifications were performed in a 50 μl reaction mixture using the TaKaRa LA TaqTM with GC Buffer kit (TAKARA BIO Inc., Shiga, Japan), following the manufacturer's instructions. Thermal cycling was 94 °C for 5′, 94 °C for 30″, 48 °C for 30″, and 70 °C for 10′ for 35 cycles; 15′ at 72 °C as a final extension. Amplified PCR products were gel purified and cloned into a pGEM-T Easy vector (Promega, Madison, WY, USA). Sequencing was performed at Macrogen Inc. (Seoul, Korea). The complete sequence of R2 was obtained through the primer walking method. A total of 48 clones were sequenced to analyze the 5′ junction of the element with the 28S gene. Sequences were edited and assembled using MEGA4 (Tamura et al., 2007).
Quantification of rDNA units and R2 copies within the T. cancriformis genome was performed through dot blot analysis. Genomic DNA was spotted onto positively charged nylon membranes (GE Healthcare Limited, Amersham Place, Little Chalfont, Buckingamshire, UK) in a series of dilutions (2000-15.6 ng); probe lanes had dilutions ranging from 5 to 0.04 ng for the 18S probe, and from 0.1 to 0.00078 ng for the R2 probe. To quantify the percentage of rDNA units, the blotted membrane was hybridized with a 400 bp long 18S probe obtained using primers 18-5′>18i (Table 2). To score the percentage of R2-inserted units, the filter was hybridized with a 1309 bp probe specific for R2 (primer pair DIN>RIN; Table 2). Hybridizations were performed under highly stringent conditions, with the final wash at 65 °C in 0.1 × SSC, 0.1% SDS. Probe labeling and blot detection were performed using the DIG High Prime DNA Labeling and Detection Starter Kit I and the CDP-Star (Roche Diagnostics GmbH, Mannheim, Germany). Images were analyzed using ImageJ (Rasband, 1997–2007).
The open reading frame was found using the ORF Finder tool (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). The phylogenetic analysis was performed on amino-acid sequences using the alignment of Kojima and Fujiwara (2005), to which all GenBank available R2-encoded proteins were added: Blattella germanica (Accession number: EF014490; Kagramanova et al., 2007), Amblyomma americanum (AY682792), Boophilus microplus (AY682793), Ixodes scapularis (AY682794), Argas monolakensis (AY682796) (Bunikis and Barbour, 2005), and Nematostella vectensis (Kojima et al., 2006). Amino-acid sequences were aligned with the MAFFT software online version (http://align.bmr.kyushu-u.ac.jp/mafft/online/server/) using the G-INS-i algorithm with BLOSUM62 matrix. Neighbor Joining and Maximum Parsimony dendrograms were computed using PAUP* 4.0b10 (Swofford, 2003), with gaps treated as informative characters; bootstrap values were obtained after 1000 replicates. The Bayesian phylogenetic tree was constructed using MrBayes 3.1 (Ronquist and Huelsenbeck, 2003). Monte Carlo Markov chains ran for 2 million generations, with trees sampled every 100 generations; the first 332 trees were discarded as burn-in. In all analyses, the SLACS element (CAA34931) of Trypanosoma brucei (Aksoy et al., 1990) was used as outgroup.
Evaluation of R2 activity
R2 activity was analyzed through 5′ truncation patterns as described in Pérez-Gonzalez and Eickbush (2001). Truncated element copies were obtained by PCR amplification (see above) using the 28S-F2 primer, annealing 62 bp upstream of the element insertion site, coupled with various R2-specific primers: RIN2, RIN3, RIN4, and RIN5 (Table 2). These primers anneal 3020, 1899, 1251, and 699 bp downstream of the insertion site, respectively. Individuals were separated into four groups consisting of 10 specimens each: 2004 females, 2004 males, 2006 females, and 2006 males. All groups were screened with all primer pairs. A total of 10 μl of each amplification product were separated on a 2% agarose gel and Southern blotted onto a positively charged nylon membrane (GE Healthcare). For every primer pair, a specific probe was designed and used to hybridize with the related PCR product: DIN>RIN2 (963 bp); DIN3>RIN3 (694 bp); DIN4>RIN4 (723 bp); DIN5>RIN5 (451 bp). Membrane detection was performed using the Roche kits mentioned above. The images were analyzed using Total Lab100 software (Nonlinear Dynamics, Ltd., Newcastle on Tyne, UK); through software evaluations, bands belonging to different individuals were considered the same truncation variant for differences up to ±10 bp.
28S rRNA sequence variation
The nucleotide variability of the 28S genes harboring or lacking R2 was analyzed through the amplification, cloning, and sequencing (as above described) of two regions extending from the R2 insertion site to 738 bp upstream and 810 bp downstream (Figure 1). The analysis was performed on eight individuals of the 2004 sample (M2, M3, M4, M6, F24, F25, F27, and F32); three of these specimens lack the R2 full-length element (M4, M6, F32). Sequences have been entered in Genbank, under the accession numbers GU220077–GU220356.
Graphic representation of the regions sequenced for the 28S analyses, with corresponding primer pairs (listed in Table 2). The primer RIN5 was specifically used for individuals M6 and M9, which lack the complete element (see text).
Proportions of nucleotide differences (calculated as mean p-distances within each individual, p-D) and gene diversities (H) were calculated for both 28S regions; each value was taken as data point for further elaboration. Two-tailed Student t-tests, with equal variance, were computed to assess the significance of differences among the scored variability values assuming that clones harboring R2 are more variable than those lacking the insertion. A second comparison was performed between clones belonging to the individuals with the complete element (M2, M3, F24, F25, and F27) and individuals without the complete element (M4, M6, and F32), both for rDNA units harboring R2 and lacking R2. Finally, a test for selection was performed on both single and pooled datasets using the Tajima's D parameter.
Results
The R2 element in T. cancriformis
To construct the complete sequence, six clones containing the whole insertion site at the 5′ terminus (5′-TTAA↓GGTAGC-3′; Burke et al., 1999; Kojima and Fujiwara, 2005) were first considered. Other clones showed deletions of the 28S gene that range from 4 to 10 bp.
The sequence of the full-length R2 element here characterized is a consensus of all sequenced fragments obtained by primer walking. It is 3583 bp long (GenBank A.N.: EU854578) and exhibits an A+T content equal to 53%. The sequencing of the R2 5′ end showed a poly-T run of five nucleotides in all analyzed clones; the analysis of the 3′ end revealed a poly-A tail, which is another common feature of R2 mobile elements (Burke et al., 1999; Kojima and Fujiwara, 2005). The R2 sequence contains a 3093 bp long ORF, located between nucleotide 177 and nucleotide 3272, coding for 1031 amino acids. As expected, the ORF is characterized by a reverse transcriptase domain and an endonuclease domain (Figure 2). Moreover, it exhibits a single zinc-finger motif. The comparison between the ORF nucleotide sequences of the R2 elements from Triops longicaudatus and T. cancriformis shows that 881 out of 1503 bp are variable sites (58%) and they occur at the first codon position (32.8%) at the second (26.6%) and at the third (40.6%). In total, 346 amino-acid substitutions are scored between the two lineages.
Graphic representation of R2 truncation variants distribution (shown by solid vertical lines) in the 40 T. cancriformis individuals. Dashed horizontal lines represent sequences missing in all elements from that individual; the dotted vertical line represents a change in the scale of the x axis. A diagram of the R2 element and the primer names/positions with respect to the insertion site are shown at the top. Pattern box indicates the zinc-finger motif; RT, retrotranscriptase domain; EN, endonuclease domain.
In T. cancriformis, R2 occurs at low copy number: only 0.54–5.3% of rDNA units (that constitute the 0.1% of the genome) have R2 insertions.
Phylogenetic analysis
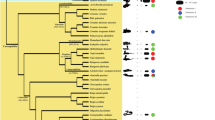
The same terminal branching pattern is observed for all phylogeny estimation methods, even if with different node support values. In the Neighbor Joining dendrogram (Figure 3), clades A, B, C, and D and subclades found by Kojima and Fujiwara (2005) can be recognized with some differences possibly occurring owing to the addition of new sequences. The main variation is given by the presence of a new subclade, named D6, that is composed by R2 elements from three tick species (family Ixodidae): R2Is (I. scapularis), R2Aa (A. americanum), and R2Bmi (B. microplus). This subclade represents the sister branch of the D5 subclade. The element of the fourth tick species (R2Amo, A. monolakensis) lies, instead, in clade A, being basal to the A2 and A3 subclades. R2 from B. germanica (R2Bg) belongs to the A2 subclade, as sister branch of R2Ha (Hasarius adansoni, jumping spider) and finally R2NvecA (N. vectensis, sea anemone) belongs to subclade D4.
The R2 sequence of T. cancriformis here characterized (henceforth called R2Tc) is in a basal position in the D5 subclade, whereas the element belonging to the congeneric T. longicaudatus (R2Tl) still lies in the A1 subclade (Kojima and Fujiwara, 2005).
Truncation analysis
The results of Southern blots on R2 truncation patterns are summarized in Table 1 and Figure 2. In the 2004 sample, a total of 318 truncations were detected, ranging from 4 to 28 per individual. In the 2006 sample, 239 variants were scored, ranging from 2 to 20 per individual. Generally speaking, a wide range of truncation profiles has been scored, as each specimen shows its own set of truncations. No ancestral variants have been detected: indeed, there is not any truncation variant present in all individuals; the most widespread one is found in 10 and 7 individuals of 2004 and 2006 collections, respectively. Moreover, six individuals, from both 2004 and 2006 samples (Table 1; Figure 2), presented a set of R2 truncations (ranging from 2 to 20), but did not have the complete element.
28S sequence variability analysis
The nucleotide variability of the 28S gene was studied using two sub-samples: the first one comprised individuals harboring both complete and truncated R2 copies (individuals M2, M3, F24, F25, and F27), whereas the second sub-sample comprised individuals only with truncation variants, but lacking the complete element (individuals M4, M6, and F32; Table 3). For each specimen, sequencing was performed for both R2-inserted and R2-uninserted 28S, upstream and downstream of the insertion site (Figure 1). From 6 to 10 sequences per individual were obtained for 28S rDNAs with or without R2 insertions, for both upstream and downstream regions.
A total of 147 28S sequences 738 bp long were obtained for the upstream region: 76 carrying the element (hence R2+) and 71 without it (hence R2−). In the R2+ dataset, 110 polymorphic sites were found resulting in 56 alleles; the mean sequence diversity within individual varies widely, from 0.0014 to 0.0081 (overall=0.0048), as well as the gene diversity, from 0.533 to 1.000 (overall=0.945; Table 3). Sequences of the R2− dataset show 93 polymorphic sites and are, on average, slightly less variable: the overall sequence variability is equal to 0.0042 (varying from 0.0011 to 0.0116) and the overall gene diversity is 0.869 (ranging from 0.533 to 1.00; Table 3). It is to be noted that the 04-F27 individual shows an R2− mean p-distance value of 0.0116, significantly higher than the population average (0.0042). Grubb's test for outliers resulted significant for that value (P<0.05); therefore, it has been excluded from subsequent analyses.
For the downstream region, 133 28S sequences 810 bp long were obtained: 63 for the R2+ dataset and 70 for the R2− one. In the former alignment, 83 sites were found variable, whereas in the latter, 95 were polymorphic. Also in this region, sequence and gene diversities vary widely: 0.0015–0.0067 and 0.800–1.000, respectively, for the R2+ dataset; 0.0012–0.0086 and 0.786–1.000, respectively, for the R2− one (Table 3). Overall, R2+ and R2− values for both parameters are almost the same: sequence variability is equal to 0.0041 for both dataset; gene diversity results 0.929 and 0.943, respectively. Interindividual sequence variabilities are reported in Supplementary Tables S1 and S2.
Student t-tests performed on both sequence and gene diversity measures did not show any significant comparison between R2+ and R2− datasets. R2 presence does not seem to interfere with the 28S homogenization process. Moreover, tests conducted between individuals carrying the complete element and those without it again did not show any significant comparison, for both R2+ and R2− datatsets and for both upstream and downstream regions.
Tajima's D test performed on single R2+ and R2− alignments, as well as for pooled datasets, rejects the neutrality hypothesis in the majority of trials, especially for the upstream region; moreover, all values, whether significant or not, are negative (Table 3).
Discussion
The R2 element from T. cancriformis well matches general features of this kind of transposable element. So, a single ORF with a central reverse transcriptase domain is present, as in type II non-LTR retrotransposons (Eickbush and Jamburuthugoda, 2008). In addition, its insertion site within the T. cancriformis 28S gene is the same as in all studied organisms (Burke et al., 1999; Kojima and Fujiwara, 2005) save ticks, which show two nucleotide substitutions within the normal target sequence (Bunikis and Barbour, 2005). In T. cancriformis, a four base deletion (TTAA) was found in R2-inserted 28S, whereas in the Drosophila genus 28S deletions are larger. On the other hand, the 3′ end of R2Tc is congruent with that of Drosophila spp., with the typical poly-A tail and the deletion of the two Gs at the insertion site (George et al., 1995; Burke et al., 1999; Pérez-Gonzalez and Eickbush, 2001). These features are determined by the target primed reversed transcription mechanism, in the phase of DNA cleavage and cDNA synthesis (Christensen et al., 2006).
R2 phylogeny
Earlier studies outlined an important aspect of R2 evolution: with only few exceptions, the R2 and host phylogenies do not overlap. Two hypotheses have been put forward to explain this pattern: in the first, vertical inheritance of the element can be followed by lineage extinction or diversification in certain groups; the second hypothesis assumes the horizontal transfer of R2 between species. In a recent survey, the former has been shown as the most likely explanation (Kojima and Fujiwara, 2005), so that the incongruence between host and R2 phylogeny can be explained, almost totally, by high rates of diversification of the element and not by horizontal transfer between species. The matter is very intriguing, because deep nodes of R2 phylogeny are consistent with structural features (number of zinc-finger motifs at the N-terminus; Kojima and Fujiwara, 2005) and, if we assume a vertical inheritance followed by diversification, some still-unknown factor should underlie this consistency. Obviously, the two hypotheses (vertical vs horizontal transmission) may not be mutually exclusive.
As expected from the occurrence of one zinc-finger motif, R2Tc falls in the D clade (Kojima and Fujiwara, 2005) and does not cluster with the T. longicaudatus element, which lies within the group of elements with three zinc-finger motifs. Actually, this is not currently verifiable because the only available T. longicaudatus R2 element is not complete. Beside structural features, the two notostracan R2 lineages are also very different both at the nucleotide (58%) and amino-acid (69%) levels. Owing to its observed high rate of retrotransposition, R2 sequences are subject to a high diversification leading to the occurrence of multiple lineages within the same species and/or elimination of some lineages owing to competition for the limited number of insertion sites (Pérez-Gonzalez and Eickbush, 2001). Moreover, R2s are ancient components of the animal genome, their presence dating back at least to the splitting of cnidarians and bilaterians (Kojima et al., 2006). The antiquity and the evolutionary dynamics of R2 may explain, therefore, the lack of correlation between its phylogeny and that of the host species. In this view, the divergence of the two tadpole shrimp R2 sequences might be the result of such an evolutionary process.
Truncation analysis
R2 truncation analyses have been first conducted on laboratory stocks of Drosophila melanogaster and D. simulans. In the former, the variants distribution has been found, to some extent, well conserved, with ancestral-truncated variants being shared by individuals both within and between isofemale lines. However, some lines of D. simulans show decidedly higher R2 activity, producing less conserved truncation profiles (Pérez-Gonzalez and Eickbush, 2001, 2002; Perez-Gonzalez et al., 2003; Zhang and Eickbush, 2005). A recent survey on natural populations of D. simulans showed a high turnover rate, each individual carrying a specific collection of R2 truncations (Zhou and Eickbush, 2009). The high incidence of R2 insertions in D. simulans is correlated with a high rate of variant elimination and a lower number of inserted 28S, explanable as due to its retrotransposition creating large deletions in adjacent rDNA units, thus eliminating a number of R2 variants (Zhang et al., 2008). The dynamics of R2 in T. cancriformis is in line with these observations as (i) individuals from the same and/or different samples show very different truncation profiles, (ii) there are not ancestral variants shared by all individuals, and (iii) the percentage of rDNA units with insertions is very low (∼0.5–5%). However, a peculiarity related to the R2 elimination occurs: six tadpole shrimps show truncated variants, but not the complete element. Therefore, an active R2 is lacking in their genomes.
In D. simulans, inserted 28S are hypothesized to be eliminated by the transposition of active R2 elements, whereas genomic turnover mechanisms tend to replace deleted rDNA units with new ones for the maintenance of the ribosomal locus functionality. This, however, also creates new niches for the R2 element, which can remain active (Eickbush and Eickbush, 2007; Zhang et al., 2008). On the other hand, as a consequence of transposition-mediated deletions, the loss of R2 variants, either complete or truncated, might be dramatic: the element copy number can be reduced to very few copies (for example, a single 28S rDNA with an insertion). Moreover, genomic turnover mechanisms acting on rDNA locus might eliminate all 28S carrying insertions. Therefore, a process such as transposition-mediated deletion, together with unequal DNA exchanges acting on the few 28S units carrying the complete R2 elements, can explain why these variants are lacking in the 15% of the T. cancriformis assayed. Once the full-length copy is deleted, new insertions cannot occur and the remaining truncations would be progressively eliminated by subsequent rounds of genomic turnover mechanisms (Dover, 2002).
The loss of the R2 element from a genome is the first step toward the extinction in a given population/species, possibly leading to unclear phylogenetic patterns (see above); it is, therefore, essential to understand how this mechanism proceeds. The elimination of R2 through the interplay between transposition-mediated deletions and genomic turnover mechanisms might give a clue to the process, but how the absence of R2 can be maintained in a population is still a further, open question. In a gonochoric population, as the studied tadpole shrimp samples are, outcrosses between individuals without the complete element and individuals carrying a functional R2 will very likely result in an offspring with active (complete) elements. This would mean that the extinction of an R2 lineage in a given population, in absence of other factors, is a very unlikely event, albeit it has been already shown (Jakubczak et al., 1991). The occurrence of some selective advantages at one particular stage of the population/species life history, leading to a preferential survival of the individuals lacking R2, can be suggested. However, we neither have any direct evidence, nor it is possible to draw similar conclusions from the datasets presented so far. Alternatively, as discussed in the earlier paragraph, the peculiar dynamics of this non-LTR retrotransposon may generate the presence of multiple R2 lineages in the same genome, competing for the limited insertion sites (Pérez-Gonzalez and Eickbush, 2001). Thus, the possibility of an R2 lineage's replacement, which would prevent the annealing of designed primers, cannot be excluded.
28S variation analysis
The presence of R2 (either truncated or not) within a 28S gene can influence the ribosomal sequence homogeneity in two ways: (i) a large insertion (kilobases long) may interfere with recombination, preventing the pairing of inserted with uninserted rDNA repeats; (ii) once R2 is inserted within a 28S sequence, the ribosomal gene becomes a pseudogene and may freely accumulate mutations. In both instances, R2 insertion can hinder concerted evolution, basically avoiding the homogenization process. It can, therefore, be expected that inserted sequences would be more variable than the uninserted ones. Here, the presence of R2 does not impact on 28S sequence and gene diversity. Indeed, comparisons between R2+ and R2− 28S genes are not significant, showing very close variability values. This is in line with data on Drosophila, in which inserted and uninserted 28S are identical: this has been explained through the rapid elimination of new insertions (Eickbush and Eickbush, 2007). Differently, 28S rRNA genes carrying the Pokey element in D. pulex accumulate more mutations than those without the insertion: the authors explain this contrasting pattern as the results of the long persistence (or even the spreading) of some Pokey insertions preventing ribosomal unit recombination and homogenization (Glass et al., 2008). The R2 turnover suggested for T. cancriformis would not allow this dynamics, as the newly transposed variants would be rapidly eliminated. As argued by Glass et al. (2008), recently generated 28S-inserted copies would be indistinguishable from those that never experienced the element insertion and, because of their quick elimination, they do not significantly alter the rDNA homogenization level.
The generally low variability observed in this analysis suggests a quite efficient process of sequence conservation and the hypothesis of neutrality has been rejected in several instances. Interestingly, the majority of significant Tajima's Ds can be observed in the region upstream of the insertion site, whereas in only few instances this has been shown in the downstream region. The downstream region here characterized is homologous to that described by Glass et al. (2008) in D. pulex in which the same, non-significant values have been observed: this may indicate that this region undergoes neutral evolution.
Generally speaking, all Tajima's D values observed here are negative, evidencing an excess of low frequency polymorphisms: this can be either the results of purifying selection (that can be expected, of course) or caused by a recent expansion of new 28S variants. Measures of gene diversity are consistent with the latter scenario as the higher values obtained are expected when there are several alleles none of which reaching very high frequency. This well reconciles with the R2 turnover observed here: as recalled in the earlier paragraph, as for any retrotransposition event large rDNA units deletions occur (Zhang et al., 2008), a compensatory replacement of new 28S variant is necessary for proper functionality. Multiple cycles of rDNA unit gains and losses would boost their turnover, leading to a quite homogeneous array (as rDNA units are very recently duplicated), but at the same time let spread several single point mutations throughout the array.
It would be interesting to investigate if, in the absence of R2, the same pattern of sequence and gene diversity can be achieved. The three individuals without the complete (=active) R2 element show no difference in comparison with those having the full-length retrotransposon; however, as they were sampled from a random mating population, it is unlikely that subsequent generations will lack the active element.
The R2/rDNA ‘interplay’ can be interpreted as a reciprocal advantage: new niches for R2, more efficient homogenization of rDNA units. Does this dynamics bring further advantage to the host? The turnover of rDNA units within the array, in which the retrotransposition occurs, may lead to a greater variance in the proportion of functional/defective 28S rRNA genes between individuals. In an evolutionary perspective, this would result in more opportunities for natural selection to operate on the host.
Accession codes
References
Aksoy S, Williams S, Chang S, Richards FF (1990). SLACS retrotransposon from Trypanosoma brucei gambiense is similar to mammalian LINEs. Nucleic Acids Res 18: 785–792.
Bunikis J, Barbour AG (2005). Ticks have R2 retrotransposons but not the consensus transposon target site of other arthropods. Insect Mol Biol 14: 465–474.
Burke WD, Eickbush DG, Xiong Y, Jakubczak J, Eickbush TH (1993). Sequence relationship of retrotransposable elements R1 and R2 within and between divergent insect species. Mol Biol Evol 10: 163–185.
Burke WD, Malik HS, Jones JP, Eickbush TH (1999). The domain structure and retrotransposition mechanism of R2 elements are conserved throughout arthropods. Mol Biol Evol 16: 502–511.
Christensen SM, Ye J, Eickbush TH (2006). RNA from the 5′ end of the R2 retrotransposon controls R2 protein binding to and cleavage of its DNA target site. Proc Natl Acad Sci USA 103: 17602–17607.
Dover GA (1982). Molecular drive, a cohesive model of species evolution. Nature 299: 111–117.
Dover GA (2002). Molecular drive. Trends Genet 18: 587–589.
Eickbush TH, Burke WD, Eickbush DG, Lathe III WC (1997). Evolution of R1 and R2 in the rDNA units of the genus Drosophila. Genetica 100: 49–61.
Eickbush TH, Eickbush DG (2007). Finely orchestrated movements: evolution of the ribosomal RNA genes. Genetics 175: 477–485.
Eickbush TH, Jamburuthugoda VK (2008). The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res 134: 221–234.
Fisher DC (1990). Rate of evolution in living fossils. In: Briggs DEG, Krauther PR (eds). Paleobiology. Blackwell Scientific: London. pp 152–159.
George JA, Burke WD, Eickbush TH (1995). Analysis of the 5′ junctions of R2 insertions with the 28S gene: implications for non-LTR retrotransposition. Genetics 142: 853–863.
Glass SK, Moszczynska A, Crease TJ (2008). The effect of transposon Pokey insertions on sequence variation in the 28S rRNA gene of Daphnia pulex. Genome 51: 988–1000.
Halanych KM (2004). The new view of animal phylogeny. Annu Rev Ecol Evol Syst 35: 229–256.
Hillis DM, Dixon MT (1991). Ribosomal DNA: molecular evolution and phylogenetic inference. Q Rev Biol 66: 411–453.
Jakubczak JL, Burke WD, Eickbush TH (1991). Retrotransposable elements R1 and R2 interrupt the rRNA genes of most insects. Proc Natl Acad Sci USA 88: 3295–3299.
Kagramanova AS, Kapelinskaya TV, Korolev AL, Mukha DV (2007). R1 and R2 retrotransposons of German cockroach Blattella germanica: a comparative study of 5′-truncated copies integrated into the genome. Mol Biol 41: 546–553.
Kojima KK, Fujiwara H (2004). Cross-genome screening of novel sequence-specific non-LTR retrotransposons: various multicopy RNA genes and microsatellites are selected as targets. Mol Biol Evol 21: 207–217.
Kojima KK, Fujiwara H (2005). Long-term inheritance of the 28S rDNA-specific retrotransposon R2. Mol Biol Evol 22: 2157–2165.
Kojima KK, Kuma K, Toh H, Fujiwara H (2006). Identification of rDNA-specific non-LTR retrotransposons in Cnidaria. Mol Biol Evol 23: 1984–1993.
Korn M, Marrone F, Pérez-Bote JL, Machado M, Cristo M, Cancela da Fonseca L et al. (2006). Sister species within the Triops cancriformis lineage (Crustacea, Notostraca). Zool Scr 35: 301–322.
Malik HS, Burke WD, Eickbush TH (1999). The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol 16: 793–805.
Mallatt JM, Garey JR, Shultz JW (2004). Ecdysozoan phylogeny and Bayesian inference: first use of nearly complete 28S and 18S rRNA gene sequences to classify the arthropods and their kin. Mol Phylogenet Evol 31: 178–191.
Mantovani B, Cesari M, Luchetti A, Scanabissi F (2008). Mitochondrial and nuclear DNA variability in the living fossil Triops cancriformis (Bosc, 1801) (Crustacea, Branchiopoda, Notostraca). Heredity 100: 496–505.
Mantovani B, Cesari M, Scanabissi F (2004). Molecular taxonomy and phylogeny of the ‘living fossil’ lineages Triops and Lepidurus (Branchiopoda: Notostraca). Zool Scr 33: 367–374.
Nei M, Rooney AP (2005). Concerted and birth-and-death evolution of multigene families. Annu Rev Genet 39: 121–152.
Penton EH, Crease TJ (2004). Evolution of the transposable element Pokey in the ribosomal DNA of species in the subgenus Daphnia (Crustacea: Cladocera). Mol Biol Evol 21: 1727–1739.
Perez-Gonzalez CE, Burke WD, Eickbush TH (2003). R1 and R2 retrotransposition and elimination from the rDNA loci of the X and Y chromosomes of Drosophila melanogaster. Genetics 165: 675–685.
Perez-Gonzalez CE, Eickbush TH (2001). Dynamics of R1 and R2 elements in the rDNA locus of Drosophila simulans. Genetics 158: 1557–1567.
Pérez-Gonzalez CE, Eickbush TH (2002). Rates of R1 and R2 retrotransposition and elimination from the rDNA locus of Drosophila melanogaster. Genetics 162: 799–811.
Rasband WS (1997–2007) ImageJ. U.S. National Institutes of Health, Bethesda, Maryland, USA, http://rsb.info.nih.gov/ij/.
Ronquist F, Huelsenbeck JP (2003). MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574.
Swofford DL (2003). PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates: Sunderland, Massachusetts.
Tamura K, Dudley J, Nei M, Kumar S (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599.
Zhang X, Eickbush TH (2005). Characterization of active R2 retrotransposition in the rDNA locus of Drosophila simulans. Genetics 170: 195–205.
Zhang X, Zhou J, Eickbush TH (2008). Rapid R2 retrotransposition leads to the loss of previously inserted copies via large deletions of the rDNA locus. Mol Biol Evol 25: 229–237.
Zhou J, Eickbush TH (2009). The pattern of R2 retrotransposon activity in natural populations of Drosophila simulans reflects the dynamic nature of the rDNA locus. PLoS Genet 5: e1000386.
Acknowledgements
We thank Dr Dani Boix, Dr Jordi Sala (Girona University), and Dr Loris Mularoni (IMIM, Barcelona) for collecting the specimens, and Professor Haruhiko Fujiwara for supplying part of the R2 protein alignment. This work was funded by Donazione Canziani and Fondazione del Monte di Bologna e Ravenna grants. We also thank two anonymous referees for their constructive criticism that substantially improved this paper. This paper is dedicated to Professor Franca Scanabissi: from her reproductive and ultrastructural point of view, she has beautifully introduced us to the Triops (and other Branchiopoda) world, her collaboration having always strongly supported the research group.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Heredity website
Supplementary information
Rights and permissions
About this article
Cite this article
Mingazzini, V., Luchetti, A. & Mantovani, B. R2 dynamics in Triops cancriformis (Bosc, 1801) (Crustacea, Branchiopoda, Notostraca): turnover rate and 28S concerted evolution. Heredity 106, 567–575 (2011). https://doi.org/10.1038/hdy.2010.86
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2010.86
Keywords
This article is cited by
-
Characterization and Evolution of Germ1, an Element that Undergoes Diminution in Lampreys (Cyclostomata: Petromyzontidae)
Journal of Molecular Evolution (2019)
-
Hybridogenesis and a potential case of R2 non-LTR retrotransposon horizontal transmission in Bacillus stick insects (Insecta Phasmida)
Scientific Reports (2017)
-
The Non-LTR Retrotransposon R2 in Termites (Insecta, Isoptera): Characterization and Dynamics
Journal of Molecular Evolution (2011)