Abstract
The Brazilian rosewood (Dalbergia nigra) is an endangered tree endemic to the central Brazilian Atlantic Forest, one of the world's most threatened biomes. The population diversity, phylogeographic structure and demographic history of this species were investigated using the variation in the chloroplast DNA (cpDNA) sequences of 185 individuals from 19 populations along the geographical range of the species. Fifteen haplotypes were detected in the analysis of 1297 bp from two non-coding sequences, trnV-trnM and trnL. We identified a strong genetic structure (FST=0.62, P<0.0001), with a latitudinal separation into three phylogeographic groups. The two northernmost groups showed evidence of having maintained historically larger populations than the southernmost group. Estimates of divergence times between these groups pointed to vicariance events in the Middle Pleistocene (ca. 350 000–780 000 years ago). The recurrence of past climatic changes in the central part of the Atlantic forest, with cycles of forest expansion and contraction, may have led to repeated vicariance events, resulting in the genetic differentiation of these groups. Based on comparisons among the populations of large reserves and small, disturbed fragments of the same phylogeographic group, we also found evidence of recent anthropogenic effects on genetic diversity. The results were also analysed with the aim of contributing to the conservation of D. nigra. We suggest that the three phylogeographic groups could be considered as three distinct management units. Based on the genetic diversity and uniqueness of the populations, we also indicate priority areas for conservation.
Similar content being viewed by others
Introduction
The Brazilian Atlantic Forest is considered the second largest tropical forest in South America, covering an area of more than one million square kilometres along the Brazilian coast and extending to eastern Paraguay and north-eastern Argentina (Oliveira-Filho and Fontes, 2000). This biome includes both evergreen forest, which runs along the coastline covering mountain chains at low and medium elevations (⩽1000 m), and semi-deciduous forest, extending across a plateau (usually >600 m) in the centre and south-eastern interior of Brazil (Morellato and Haddad, 2000; Oliveira-Filho and Fontes, 2000). It is one of the most diverse biomes in the world, with high species richness and endemisms, and is considered one of the most threatened ecosystems on Earth due to intense disturbance, having been reduced to only 7.5% of its original area (Myers et al., 2000).
Despite an increase in research efforts in the past few years, the knowledge about the evolutionary history of the Atlantic Forest is still limited. Phylogeographical studies have shown that the Atlantic Forest has a complex history that resulted in large lineage divergences. North–south genetic discontinuities in the central part of the Atlantic Forest (from the Bahia to São Paulo states) were found in some animal species, such as small mammals (Metachirus nudicaudatus; Costa, 2003), lizards (Gymnodactilus darwin, Pellegrino et al., 2005), sloths (Bradypus variegates; Moraes-Barros et al., 2006), birds (Xiphorhynchus fuscus; Cabanne et al., 2007) and canids (Cerdocyon thous; Tchaicka et al., 2007). The genetic split found in these studies is located in a region between the Jequitinhonha and Doce Rivers (see Figure 1 for river locations). The separation of these lineages has been dated in some studies to the Pleistocene (Cabanne et al., 2007; Tchaicka et al., 2007) and even before the Pleistocene (Costa, 2003; Pellegrino et al., 2005). The few phylogeographic studies in plants have also shown genetic discontinuities in the central part of the Atlantic Forest, but the location of these splits are more southern (southern Espirito Santo/northern Rio de Janeiro states, ∼21°S), as in the legume trees (Caesalpinia echinata, Lira et al., 2003; Hymenaea courbaril, Ramos et al., 2009). However, in conifer trees (Podocarpus sellowi, P. brasiliensis and P. lambertii, Ledru et al., 2007) the split was observed at more northern locations (15°S). Although the studies in plants have attributed these genetic splits to climatic changes of the Quaternary, none has estimated the time since splitting. In addition, these phylogeographic studies with plants had a sparse sampling through the central Atlantic forest. Additional phylogeographic studies would help to reconstruct the evolutionary history of the central Atlantic Forest, improving conservation and management measures such as the identification of priority populations/areas for reserve implementation.
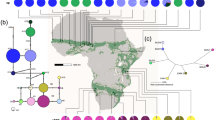
Map showing the geographical distribution of the 19 sampled populations for the phylogeographic study of Dalbergia nigra (black points) and the original distribution of the Brazilian Atlantic Forest in South America (grey). The approximate distribution limits of D. nigra are indicated by broken lines covering five Brazilian States: Bahia (BA), Minas Gerais (MG), Espírito Santo (ES), Rio de Janeiro (RJ) and São Paulo (SP). See Table 1 for the population abbreviations and other details.
Vegetation changes in the Atlantic Forest related to climatic changes during the Pleistocene have been registered in paleopalinological studies, with the replacement of large areas of forests by subtropical grasslands and savannas during cooler and drier conditions (Behling 1995, 2002; Behling and Negrelle, 2001). Based on paleoclimatic models, Carnaval and Moritz (2008) predicted the presence of historically stable areas (refugia) in the Atlantic Forest during the Late Quaternary, one large area in the central region (the Bahia refugium) and another smaller area (the Pernambuco refugium). These match the current centres of the endemism of several taxa and published phylogeographic studies based on animal mitochondrial DNA (mtDNA).
In addition to past historical events, it is known that human-mediated fragmentation and deforestation can also promote alterations in the diversity and genetic structure of species, frequently depleting genetic diversity within populations and increasing genetic differentiation among populations (Young et al., 1996; Aguilar et al., 2008). The high deforestation rate of the Atlantic Forest could lead to a loss of the signature of historical processes behind the genetic structure of species (Carnaval et al., 2009). Thus, studies that take into account both of the factors that affect the current genetic structure of species—historical and human-mediated events—are required to better understand the evolutionary process and establish effective conservation measures.
Here, we analyse the phylogeography of Dalbergia nigra (Vell.) Allemao ex Benth (Papilionoideae), known as the Brazilian rosewood or jacarandá-da-Bahia, one of the most valuable tree species endemic to the central Atlantic Forest, which occurs from Bahia to the northern São Paulo States (Carvalho, 1994). This species produces a high-quality wood that is highly valued for the manufacture of musical instruments and fine furniture, thus resulting in its overcutting since the colonization of Brazil (1500 AD) (Carvalho, 1994). D. nigra is a threatened tree that is in the vulnerable category due to its over-exploration, the absence of replacement plantations and the deforestation of the Atlantic Forest (IUCN, 2008). D. nigra is extremely rare in nature, and its international trade has been prohibited since the 1990s by the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES, 2008).
We performed a dense sampling throughout the distribution range of D. nigra to obtain a more geographically detailed understanding of the evolutionary history of this endemic species of the central Atlantic Forest. Using chloroplast DNA (cpDNA) sequences, the phylogeographic structure and demographic history were investigated. In addition to classical phylogeographic methodologies, we performed a demographical analysis using the ‘isolation with migration’ model (IM, Hey and Nielsen, 2004, 2007), which is based on coalescent theory. This methodology provides estimates of the effective population sizes, time since the populations split and rate of migration after a population split even using a single locus (see Qiu et al., 2009). This study was conducted to answer the following two questions: (1) What is the influence of the historical events of the Pleistocene on the current genetic diversity and structure of D. nigra, and specifically, are there genetic disjunctions along its distribution? (2) In addition to historical events, were the genetic diversities of populations affected by human-mediated fragmentation? The existence of a well-defined phylogeographic structure could be informative for understanding historical climatic events. A lower diversity in small forest fragments in relation to great reserve areas, both located in the same phylogeographic group, could be associated with more recent events, such as human-mediated fragmentation. Although cpDNA is not as powerful for detecting alterations in genetic structure in fragmented populations as more polymorphic markers, it can be expected that pronounced fragmentation results in a decrease in the genetic diversity of small populations, which could also be detected using cpDNA. All of the results obtained here were used to suggest conservation strategies for this endemic and threatened species of the Atlantic Forest.
Materials and methods
Study species and population sampling
Dalbergia nigra trees reach 15–25 m in height and 40–80 cm in diameter at breast height (Carvalho, 1994). It has a short flowering period (November–December) and a long fruiting period (January–September), both in supra-annual and irregular phenological cycles (data from southeast Brazil; Engel and Martins, 2005). D. nigra is pollinated by bees (personal observation), and its seeds are dispersed by the wind (Carvalho, 1994). Its mating system is not known, but the congeneric tree D. miscolobium, from the Brazilian Cerrado exhibits outcrossing with an apparent self-incompatibility system (Gibbs and Sassaki, 1998), which possibly suggests a similar mating system in D. nigra.
Nineteen populations of D. nigra were sampled, covering most of its current range in the Atlantic Forest (13°–23°S and 39°–45°W), at altitudes ranging from 58 to 870 m (Figure 1; Table 1). The populations were located in four Brazilian States: Bahia, Minas Gerais, Espirito Santo and Rio de Janeiro. The distance among the D. nigra populations sampled in this study ranged from approximately 8 km (SCR and DIO) to 1166 km (AMA and AIA) (Figure 1). Two populations (RDO and VRD) were located in large, well-preserved biological reserves (36 000 and 22 000 ha, respectively), and one (SCR) was in a small private reserve (51 ha). The other populations were located in small, disturbed forest fragments along highways or inside farms and covered areas smaller than 10 ha, except for population DIO (40 ha). Leaves were collected from about 10 adult trees in each population, totalling 185 individuals. These trees were 8–25 m tall. To ensure adequate coverage of each studied population, random samples were taken from trees at a 10- to 600-m distance from one another across each population. Following collection, the leaves were shipped in ice from the field to the laboratory and were thereafter stored at −70 °C until the DNA extraction was performed.
DNA amplification and sequencing
Total DNA was extracted from the ground leaves and stored at −20 °C until further analyses. The screening for cpDNA amplification and polymorphism in D. nigra was conducted on a subset of individuals using 15 universal primer pairs for non-coding cpDNA regions previously described: trnL-trnF, trnL intron, trnT-trnL (Taberlet et al., 1991); trnV-trnM, psaA-trnS, trnC-trnD, trnH-trnK, trnK1-trnK2, psbC-trnS (Demesure et al., 1995); trnH-psbA, psbB-psbF, rpl20-rps12, trnS-trnG (Hamilton, 1999); trnQ-trnS (Grivet et al., 2001); ccmp4 L-atpH (Weising and Gardner, 1999). Two non-coding cpDNA regions, the trnV-trnM intergenic spacer and trnL intron, revealed polymorphisms in the analysed individuals and were therefore selected for a large-scale survey of haplotype variation in D. nigra.
Polymerase chain reaction (PCR) was performed in a 25-μl total volume containing 10–20 ng of genomic DNA, 1X Taq buffer with 2.0 mM MgCl2, 0.2 ng of BSA (bovine serum albumin), 200 μM of each dNTP, 0.5 μM of each primer, 1U Taq DNA polymerase (Phoneutria, Belo Horizonte, Brazil) and autoclaved deionized water. The amplifications were carried out in an Eppendorf thermocycler with an initial step at 94 °C for 4 min (trnV-trnM) or 2 min (trnL), followed by 35 cycles at 94 °C for 1 min, 59 °C (trnV-trnM) or 56 °C (trnL) for 1 min, and 72 °C for 2 min (trnV-trnM) or 1 min (trnL), plus a final extension at 72 °C for 7 min. Following the PCR, the products were viewed on 1% TAE agarose gels and purified by 20% PEG (polyethylene glycol) precipitation. Each PCR product was then double-strand sequenced using the DYEnamic ET dye terminator sequencing kit (GE Healthcare, Buckinghamshire, England, UK). Sequencing reactions were analysed on a MegaBACE 1000 automated sequencer (GE Healthcare). To determine any PCR errors or sequencing artefacts, two or more independent PCR amplifications were sequenced for samples that exhibited an unexpected polymorphism.
The quality of the cpDNA sequences was checked and overlapping fragments were assembled using PHRED v. 0.20425 (Ewing and Green, 1998; Ewing et al., 1998), PHRAP v. 0.990319 (http://www.phrap.org/) and CONSED 12.0 (Gordon et al., 1998) to produce high quality consensus sequences. These sequences were aligned using CLUSTAL-W (Thompson et al., 1994) implemented in the MEGA 3 program (Kumar et al., 2004), with default gap penalties and manual verification. The trnV-trnM and intron trnL sequences were deposited in GenBank under accession numbers GU816029-GU816038 and GU816025-GU816028, respectively. All analyses in this study were performed using the combined data from the cpDNA sequences.
Population and phylogeographical analyses
The cpDNA haplotypes were defined by analysing sequences with DNASP 4.01 (Rozas et al., 2003). The haplotype diversity (h) and nucleotide diversity (π) were estimated in ARLEQUIN 3.01 (Excoffier et al., 2005). The allelic (haplotypic) richness (Hr) was calculated using the rarefaction method, standardized for the lower sample size, implemented in CONTRIB 1.02 (Petit et al., 1998) (software available at http://www.pierroton.inra.fr/genetics/labo/Software/Contrib/). The paired t-test for independent samples implemented in BioEstat 5.0 (Ayres et al., 2007) was used to compare the genetic diversity (h, π and Hr) of small fragments with large reserve areas. The contributions of each population to the total haplotypic diversity (CT) and total haplotypic richness (CTR) were estimated using CONTRIB and partitioned into two components, the contribution due to a population's own level of diversity (CS and CRS), and its differentiation from other populations (CD and CRD), respectively (Petit et al., 1998). To investigate the coefficient of genetic differentiation across all populations, two analyses were performed in HAPSTEP (update 2001; Pons and Petit, 1996): the GST value based only on the haplotype frequencies and the NST value influenced by both the haplotype frequencies and distances among haplotypes (number of mutational steps). The existence of phylogeographic structure is suggested when NST is significantly higher than GST (Pons and Petit, 1996). The phylogenetic relationships among the haplotypes were estimated using the median-joining algorithm implemented in NETWORK 4.1 (Bandelt et al., 1999).
An analysis of molecular variance (AMOVA) using the pairwise difference was performed in ARLEQUIN to investigate the population structure (FST values) in D. nigra. To define alternative patterns of population subdivision and to estimate possible barriers to gene flow, we used, respectively, a spatial analysis of molecular variance (SAMOVA) based on the pairwise difference, performed in SAMOVA 1.0 (Dupanloup et al., 2002), and the Monmonier maximum difference algorithm (Monmonier, 1973), performed in BARRIER 2.2 (Manni and Guérard, 2004). Using a simulated annealing approach, the SAMOVA program identifies groups of populations (k, which ranged from two to eight) that are geographically homogenous and maximally differentiated from each other (higher FCT values) with 1000 permutations. By contrast, the BARRIER program implements Monmonier's algorithm, which identifies possible genetic barriers among the most differentiated groups of populations, spatially clustered, creating a Delaunay triangulation network to connect adjacent populations and, consequently, a Voronoi tessellation set (Manni et al., 2004).
A multiple regression analysis (Smouse et al., 1986) was performed using 10 000 permutations in FSTAT version 2.9.3.2 (Goudet, 2002). This analysis tests the effect of the independent variables, linear geographic distance between population pairs and their geographic location, on the average corrected genetic distances among individuals from population pairs. The first variable reflects isolation by distance, whereas the second suggests vicariance events as the principal process of genetic divergence. The linear geographic distance matrix was inferred in kilometres separating population pairs, whereas the geographical location matrix was constructed by assigning a value of zero to population pairs within the same group (as defined in SAMOVA) and one to those in different groups. The genetic distances between the population pairs were estimated under the Kimura-2 parameter model using MEGA.
Demographic analyses and divergence time estimates
The hypothesis of population expansion was tested using different approaches. The pairwise mismatch distribution (Rogers and Harpending, 1992) was estimated in ARLEQUIN. Two groups of neutrality tests were computed in DNASP and ARLEQUIN: (1) Tajima's D (Tajima, 1989, Fu and Li's (1993) F* and D* considering the frequency of mutation (segregating sites); and (2) Fu's (1997) FS based on the haplotype distribution. The demographic history of a population could be inferred by comparing such neutrality tests, given that a range expansion is suggested when Tajima's D and Fu's FS are significantly negative, but F* and D* are not (Fu, 1997). The maximum likelihood coalescence method implemented in LAMARC 2.1.2 (Kuhner, 2006) was used to estimate the population growth rate (g) under the exponential growth model. The analysis was conducted using the Markov Chain Monte Carlo (MCMC) sampling algorithm with 10 initial chains with 10 000 sampled genealogies each and two final chains with 200 000 sampled genealogies each.
The isolation with migration (IM) coalescent model, as implemented in the program IM (update 2007; Hey and Nielsen, 2004), was used to estimate the divergence times (t) among the groups of populations of D. nigra previously defined by SAMOVA. Moreover, the program IM also estimated the following parameters: two migration rate (m1 and m2) parameters and the effective population sizes of the current (θ1 and θ2) and ancestral populations (θA). Several runs of IM were first performed to determine the appropriate previous values that captured completely the posterior distributions of the parameters. Subsequently, the runs were processed to verify the convergence of the selected previous parameters with different random seed numbers, as assessed by the autocorrelation of the parameters lower than 0.01, effective sample sizes (ESS) larger than 100 and update rates higher than 2%. The previous values used for the final analyses were as follows: t=15, θ1=θ2=θA=15 and m1=m2=5. Two final runs were conducted with four Metropolis-coupled MCMC, each run for 20 million steps after a burn-in period of 5 million steps. The MCMC approach assumed the HKY model of sequence evolution and an inheritance scalar of 0.5 for the maternally inherited cpDNA. The time units were converted to an estimate of the absolute time, T (time in years), using T=t/uk, where u is the number of substitutions per site per year and k is the sequence length of the cpDNA region under study. Considering that a substitution rate had not yet been estimated for the cpDNA genome of Dalbergia, we used the cpDNA synonymous substitution rate of 1.01 × 10−9 per site per year found in seed plants (Graur and Li, 1999).
In addition to the divergence time estimated using the coalescent-based approach, the timing of the divergences among the groups of populations was also estimated as T=dA/2 μ, where dA was the net pairwise divergence per base pair, as obtained in MEGA 3 under the Kimura-2 model, and μ was the nucleotide substitution rate per site (Nei and Kumar, 2000). The substitution rate was the same as in the analyses outlined above. In addition to reporting the point estimate for dA, a conservative range for divergence time was calculated by applying the 95% confidence interval (CI±1.98 s.e.).
Results
Diversity and phylogeographic structure
A total of 1297 bp from two cpDNA regions (trnV-trnM intergenic spacer—717 bp, and trnL intron—580 bp) of 185 individuals of D. nigra were sequenced. The molecular diversity indexes are shown in Table 1. These cpDNA sequences represented 15 haplotypes with 11 polymorphic sites, including one transition, nine transversions and only one indel (Table 2). The higher number of transversions than transitions observed in the analysed sequences was expected because the cpDNA regions have a higher AT content, and thus a significant tendency towards transversions in the nucleotides flanked by A and/or T (Kelchner, 2000).
The haplotype diversity (h) for each population ranged from 0 to 0.736, the nucleotide diversity (π) from 0 to 0.00084 and the haplotypic richness (Hr) from 0 to 2.222 (Table 1). The total haplotype and nucleotide diversities were 0.7525 (s.d.±0.0211) and 0.00089 (s.d.±0.00064), respectively. Four of the 19 analysed populations were monomorphic (ITA, EUN, DIO and SMA), whereas the remaining populations showed two to five haplotypes (Tables 1 and 2; Figure 2). Haplotypes H2, H6, H13 and H15 were unique to populations AMA, MED, VNI and AIA, respectively (Table 2).
Map showing the geographic distribution of the cpDNA haplotypes in the Dalbergia nigra populations. The haplotypes are identified in the legend. The broken lines indicate the genetic barriers as defined by the Monmonier algorithm. See Table 1 for the population abbreviations and molecular diversity indexes. The median-joining network analysis is represented on the upper left. The size of each circle is proportional to that haplotype frequency across populations. Each line between the haplotypes represents a mutational step (see Table 2 for details). The dotted lines indicate independent mutation events converging on a shared haplotype. The colors of circles indicate the occurrence of these haplotypes in the three population groups (black: NG group; white: SG1 group; grey: SG2 group).
The populations with the highest diversities as evaluated by h, π and Hr were those from large biological reserves (VRD and RDO) and population PPA, located in one small, disturbed fragment on a highway edge in northeastern Minas Gerais State (Table 1). The D. nigra populations contributed differently to the total genetic diversity and differentiation inferred for this species. The contributions from the diversity (CS and CRS) and differentiation (CD and CRD) components of each D. nigra population are shown in Figure 3. The highest CS and CRS values were identified in populations RDO, VRD and PPA due to their higher h and Hr values (Table 1; Figure 3). By contrast, the highest CD and CRD were seen in populations SMA, AIA and MED (Figure 3). These contributions to genetic differentiation were probably due to exclusive mutations found in medium to high frequencies in these three populations (see haplotypes H6, H14 and H15 in Table 2). The comparison of the genetic diversities of populations from small fragments (TOT, SCR, DIO, ACA, PMI and VNI) with those from large reserve areas (RDO and VRD; Figure 2), both pertaining to same geographical group (see below the results of SAMOVA program), showed significantly lower genetic diversities in all estimates in the small fragments, such as h (t=6.57, P<0.001), π (t=8.57, P<0.001) and Hr (t=8.13, P<0.001).
The contribution to the total haplotype diversity (CT) and haplotypic richness (CTR) of each population of Dalbergia nigra using the cpDNA haplotypes. The grey and black bars represent the contribution of diversity (CS and CSR) and differentiation (CD and CDR), respectively. See Table 1 for the population abbreviations.
The AMOVA analysis showed a very strong differentiation among all D. nigra populations (FST=0.624, P<0.0001). In the SAMOVA analysis, the best grouping scheme divided the distribution range of D. nigra into three main geographical groups (k=3 and FCT=0.668). When k=2 or k⩾4, the SAMOVA showed similar or lower FCT values. Furthermore, the grouping structure disappeared when k⩾4, meaning that the fourth and following groups comprised only one population. Considering these results, the Monmonier algorithm (BARRIER program) suggested the following main barriers to gene flow in the D. nigra distribution range: (1) one barrier located approximately in the northeast of Minas Gerais and south of Bahia, dividing the natural distribution of this species into northern and southern groups, and (2) another barrier located between Espirito Santo and Rio de Janeiro, thus isolating the D. nigra populations located in the extreme south of the sampling area (Figure 2). The northern group (NG) is composed of populations from Bahia and northeast Minas Gerais (13°–18°S). The southern group (SG1) is formed by populations from Espirito Santo and central and south Minas Gerais (18°–21°S). The extreme southern group (SG2) includes only populations from Rio de Janeiro (22°–23°S).
The multiple regression analysis indicated that approximately 69% of the variation (R-squared) in the genetic distances could be predicted by the geographical location (partial correlation coefficient=0.463, P<0.01) and the linear geographical distance among locality pairs (partial correlation coefficient=0.689, P<0.01). This result indicates that both vicariance processes and isolation by distance explain the present-day genetic structure of D. nigra. The NST (0.649±0.0624; P<0.05) was significantly higher than the GST (0.559±0.0690), indicating a phylogeographic structure, that is, closely related haplotypes in the D. nigra populations were found more often in the same area than less closely related haplotypes.
The phylogenetic relationships among the haplotypes were resolved by a network analysis (Figure 2). Haplotypes H1 to H6, H7 to H13 and H14 to H15 were restricted to the NG, SG1 and SG2 groups, respectively, except haplotype H7, which occurred in four individuals from population PPA of the NG group (Figure 2; Table 2). Considering that several haplotypes were connected by one mutation to one of the two most common and widespread haplotypes (H1 and H7), this star-shaped network could indicate a relatively recent population expansion in the D. nigra. Independent mutation events converging to a shared haplotype could explain some of the connections observed in the network analysis.
Demographic history and divergence time
The historical demography was analysed on the populations groups suggested by SAMOVA. The Fu's FS tests performed on the D. nigra populations were significantly negative for all populations considered together (P<0.01) and for each group considered separately (P<0.05), except SG2 (Table 3). According to Fu (1997), using a 5% cutoff in FS test will increase possibility of rejecting the null hypothesis (neutrality-demographic stability) when it is true. Thus, a clear signal of demographic expansion by Fu's FS test was revealed only for all populations together. Tajima's D tests were negative for all groups, although not significant, and the other neutrality tests (Fu and Li's D and F) were also not significant in any sample (P>0.05) (Table 3). The mismatch distribution was unimodal taking all populations into account but not for each group taken separately (Table 3). The growth rates (average g values) were high for both NG and SG1 (Table 3), however the confidence intervals were broad and had zero overlap in some of the Bayesian runs (Table 3). The g value was not estimated for the SG2 population group due to its small sample size.
The divergence time inferred from net pairwise divergence (dA=0.00071) and substitution rate (1.01 × 10−9 per site per year) suggested that NG and SG1 diverged approximately 351 000 years before the present (ybp) (CI=−323 092–1 025 072). The time of divergence between SG1 and SG2 (dA=0.00079; T=389 000 ybp; CI=−400 538–1 178 953) was similar to that between NG and SG1.
Using the IM coalescent model, the parameters estimated were coincident in two independent runs (Table 4), so the highest residence time (Hipt) estimates obtained in the run with the largest ESS for divergence time are described below. The divergence time between the main groups, NG and SG1, was estimated at 612 595 ybp (lower bound of HPD 90% interval: 372 137). The two groups of southern region, SG1 and SG2, diverged earlier, at 784 351 ybp (lower bound of HPD 90% interval: 761 450). Unlike all of the other estimates, the divergence time exhibited in all runs had update rates lower or equal to 1% and posterior distribution was always flat, that is, non-zero at the upper end, suggesting that these results should be taken with caution. The effective population size estimated was twice as large in the NG group (θNG: 2.1165; HPD 90% interval: 0.6423–4.7699) than in the SG1 group (θSG1: 1.0424; 0.2422–3.0009), whereas both were higher than the ancestral population size (θA: 0.3264; lower bound of HPD 90% interval: 0.0105), revealing a population expansion after the split of the NG and SG1 groups. The analysis between SG1 and SG2 showed a similar SG1 effective population size (θSG1: 1.2528; 0.3066–3.5307) to that obtained through the comparison between NG and SG1, indicating the robustness of the analysis. The effective population size was much lower in the SG2 (θSG2: 0.1314; 0.0263–1.7084) and the ancestral population to SG1 and SG2 (θA: 0.0263; lower bound of 90% HPD: 0.0088) than in the SG1 group. However, though the θA estimates exhibited high update rates and ESS, the posterior distributions were flat. The migration rate from NG to SG was 0.4575 (0.0025–2.0175) and the reciprocal migration was very low (0.0025; 0.0025–1.2925). The migration rate estimates between SG1 to SG2 were also low (0.0025; 0.0025–1.3775) whereas the migration from SG2 to SG1 was higher (0.1225; 0.0025–3.5725) (Table 4).
Discussion
Our analyses of the cpDNA sequences from D. nigra populations indicated a high level of genetic structure, with 62% of the total genetic diversity due to differences among populations (FST=0.624), identified as three phylogeographical groups, the northern (NG), southern 1 (SG1) and southern 2 (SG2) groups (Figure 2). D. nigra is more genetically structured than Eugenia uniflora (FST=0.21), an Atlantic Forest tree previously analysed by cpDNA PCR–RFLP (Salgueiro et al., 2004). However, the genetic structure of D. nigra is weaker than that of Caesalpinia echinata (ΦST=0.911; Lira et al., 2003), based on chloroplast microsatellites, but similar to Hymenaea courbaril, based on cpDNA sequences (FST=0.604; Ramos et al., 2009), both of which are leguminous tree species found in the Atlantic Forest. The genetic split separating the NG and SG1 groups of D. nigra was found around 18oS latitude, between the Jequitinhonha and Doce rivers, coinciding with the split zone range found in several animal species (Costa, 2003; Pellegrino et al., 2005; Moraes-Barros et al., 2006; Cabanne et al., 2007; Tchaicka et al., 2007). The other genetic disjunction in D. nigra, between the SG1 and SG2 groups, was located around 21°S, in the same region of separation between genetic groups found in another two legume trees, Caesalpinia echinata (Lira et al., 2003) and Hymenaea courbaril (Ramos et al., 2009). However, in the Podocarpus species (Ledru et al., 2007), the separation among the three groups appears not to coincide with the split zones found in legume trees. These latitudinal genetic disjunctions found in different groups of species point to historical vicariance events in the central Atlantic Forest. This scenario is corroborated by our regression analysis, which indicated that a major part of the diversity of D. nigra is due to these vicariance events (partial correlation coefficient=0.463, P<0.01).
Considering only the genetic divergences among the groups, the estimated divergence times between the NG and SG1 groups and between the SG1 and SG2 groups were similar (351 000 and 389 000 ybp, respectively). We applied the IM analysis taking into account demographic events because of possible differences in the effective population sizes between populations and gene flow after split. This analysis showed a more ancient separation time between these groups, 612 000 ybp (NG/SG1 groups) and 784 000 ybp (SG1/SG2). In spite of the differences between the estimates with the two approaches, both are concordant in pointing to vicariance events in the Middle Pleistocene. The divergence time between the NG and SG1 groups of D. nigra was similar to that found between the northern and southern groups of Cerdocyon thous, a widely distributed South American canid species (ca. 400 000–600 000 ybp; Tchaicka et al., 2007). However, the time of split between the lineages of the same latitudinal range in other studies with animal species is variable, ranging from the Late Pleistocene (Cabanne et al., 2007) to before the Pleistocene (Costa, 2003; Pellegrino et al., 2005).
Palynological data revealed that southeastern Brazil was cooler and drier during the glacial periods than it is in the present day (Behling, 2002). During the Last Glacial Maximum (LGM), around 27 500–14 500 14C years ago, the southern portion of the semi-deciduous forests was replaced by subtropical grasslands, which were displaced at least 750 km further north, from latitudes of 28°/27°S to about 20°S (Behling, 1998). These data showed that the drier conditions of the LGM caused the Atlantic Forest to become much smaller and more fragmented than it is today (Behling, 2002). As a result of this constriction, the rain forest was probably partly replaced by semi-deciduous forest in southeastern Brazil and by semi-deciduous forest and caatinga (tropical semi-desert shrub vegetation) in northeast Brazil (Behling, 1998, 2002). A palynological study in the region of the split region between the Doce and Jequitinhonha rivers, performed by Behling (1995) in Lago do Pires (17° 59′S, 42° 13′W) showed that this region in the early Holocene was dominated by species from Cerrado (the Brazilian savannah), suggesting the occurrence of long dry seasons. This study also suggested that the current dense and closed semi-decidous forest in this region was only established in the latest Holocene (beginning 970 years ago). The recurrence of climatic changes during the Pleistocene in the central part of the Atlantic forest, with cycles of forest expansion and contraction, likely led to repeated vicariance events and thus resulted in the genetic differentiation of the three population groups of D. nigra. In fact, the location of the current NG group is included in the putative Bahia refugium, a large stable region from the Doce River northward to the southern border of the São Francisco River described by Carnaval and Moritz (2008).
Our analysis showed evidence of weak population expansion in the NG and SG1 of D. nigra, as indicated by the star-like topology of the network, a borderline significance of neutrality deviation in Fu's Fs test, and the Tajima's D test showed a negative deviation, although not significant. The demographic parameters estimated in the IM corroborate with the hypothesis of population expansion because that effective sizes of the ancestral populations of the groups were lower than respective derivate populations, that is, after the split there were population expansions. The NG and SG1 exhibited high diversities (Table 3), whereas the southernmost group (SG2) had low diversity, as expected for a recently founded population. Furthermore, the NG and SG1 showed higher effective population sizes than the SG2. Because the effective size is a harmonic average of the effective size across generations, it can be suggested that the northernmost populations (NG and SG1) maintained large populations during a great period and that the SG2 may have undergone a more pronounced bottleneck. This interpretation is concordant with the indication that forest fragmentation was stronger in the southern part of Atlantic Forest as a consequence of past climatic oscillations (Behling, 1998). Population expansion in SG2 may have not been detected due to a low statistical power of the analysis, as a consequence of the small sample sizes coupled with very low levels of diversity in this group. The northern group's PPA population exhibited haplotypes typical of this group (H1, H4 and H5) and also the haplotype H7, the most common haplotype in the southern (SG1) group, suggesting secondary contact due to the recent migration of propagules from the latter group. The PPA population is located near the studied site by Behling (1995); Lago Pires, where a more recent expansion of the Atlantic semidecidous forest was registered.
In addition to the signature of the effects of past climatic changes on the phylogeographic structure of D. nigra, our data suggest that recent human-mediated activities such as deforestation and fragmentation have also affected the genetic diversity of the populations. It is noteworthy that in the same phylogeographic group (SG1), two populations located in large biological reserves (RDO and VRD) showed significantly higher levels of cpDNA diversity than populations from small forest fragments, for all measured parameters, including the allelic richness corrected for the sample size. These results are in accordance with a previous study (Ribeiro et al., 2005), which suggested that anthropogenic disturbance may have decreased the allozyme genetic diversity of D. nigra in these fragments. Disturbances may have eliminated some haplotypes that were once present in these populations. These molecular data are evidence that timber exploration and deforestation have promoted genetic depletion in this species, and also that the present diversity may represent only a part of the genetic diversity seen in the past in this wide geographical area of the Atlantic Forest. This scenario found for D. nigra could probably be extended to other timber species of the Atlantic Forest.
Implications for conservation
Phylogeography and genetic diversity data can contribute to the development of effective conservation strategies. The genetic data obtained here for D. nigra were based on chloroplast DNA diversity. Nuclear markers could result in an alteration in phylogeographical patterns obtained here. However, considering that D. nigra is a threatened species from one of the global hotspots for biodiversity conservation (Myers et al., 2000), and taking into account the precautionary principle (Myers, 1993), we consider it important to use the cpDNA data to inform conservation procedures for this species.
The criteria for the selection of priority populations to be conserved must include both the uniqueness of a population and its diversity level, especially in terms of its allelic composition (Petit et al., 1998). Considering the criteria for the recognition of management units as populations with a significant divergence of allele frequencies at nuclear or organelle loci (MU; Moritz, 1994), we suggest that the three phylogeographic groups of D. nigra could be treated as different MUs. Therefore, significant well-preserved forest remnants containing representative genetic diversity should be identified in each of the three groups to create units of conservation. The populations RDO and VRD, located in large conservation units in the SG1 group, are among those that most contributed to the total diversity of D. nigra. This fact shows the importance of preserving large areas to maintain genetic diversity. These genetic data, highlighting the uniqueness of populations in each phylogeographic group must also be considered upon sampling seeds for the ex situ conservation of D. nigra.
Special attention should be given to the northeast Minas Gerais State. In a small disturbed remnant on a highway edge in this region is located the PPA population with a high genetic diversity and containing haplotypes from the NG and SG1, with signals of current gene flow. In addition, the neighbouring population MED, also located in a small disturbed forest fragment, gave a significant contribution to the genetic diversity of D. nigra due to an exclusive mutation. There are no relevant conservation units in this region. The high genetic diversity and uniqueness found in these populations of D. nigra, plus evidence from animal studies (Ditchfield, 2000; Costa, 2003; Moraes-Barros et al., 2006; Tchaicka et al., 2007), indicate the high priority that should be given to implementing conservation units in northeast Minas Gerais.
Accession codes
References
Aguilar R, Quesada M, Ashworth L, Herrerias-Diego Y, Lobo J (2008). Genetic consequences of habitat fragmentation in plant populations: susceptible signals in plant traits and methodological approaches. Mol Ecol 17: 5177–5188.
Ayres M, Ayres Jr M, Ayres DL, Santos AAS (2007). BioEstat. Versão 5. Sociedade Civil Mamirauá, MCT—CNPq, Belém: Pará, Brasil.
Bandelt HJ, Forster P, Röhl A (1999). Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16: 37–48.
Behling H (1995). A high resolution Holocene pollen record from Lago do Pires, SE Brazil: vegetation, climate and fire history. J Paleolimnology 14: 253–268.
Behling H (1998). Late Quaternary vegetational and climatic changes in Brazil. Rev Paleobot Palynol 99: 143–156.
Behling H (2002). South and southeast Brazilian grasslands during late Quaternary times: a synthesis. Palaeogeogra Palaeoclimatol Palaeoecol 177: 19–27.
Behling H, Negrelle RRB (2001). Late Quaternary tropical rain forest and climate dynamics from the Atlantic lowland in southern Brazil. Quat Res 56: 383–389.
Cabanne GS, Santos FR, Miyaki CY (2007). Phylogeography of Xiphorhynchus fuscus (Passeriformes, Dendrocolaptidae): vicariance and recent demographic expansion in southern Atlantic forest. Biol J Linn Soc 91: 73–84.
Carnaval AC, Hickerson MJ, Haddad CFB, Rodrigues MT, Moritz C (2009). Stability predicts genetic diversity in the Brazilian Atlantic Forest Hotspot. Science 323: 785–789.
Carnaval AC, Moritz C (2008). Historical climate modelling predicts patterns of current biodiversity in the Brazilian Atlantic Forest. J Biogeogr 35: 1187–1201.
Carvalho PER (1994). Espécies Florestais Brasileiras: Recomendações Silviculturais, Potencialidades e Uso da Madeira. EMBRAPA-CNPF/SPI: Brasília.
CITES (2008). CITES: Appendix I, II and III to the Convention on International Trade in Endangered Species of Wild Fauna and Flora. US Fish and Wildlife Service: Washington.
Costa LP (2003). The historical bridge between the Amazon and the Atlantic Forest of Brazil: a study of molecular phylogeography with small mammals. J Biogeogr 30: 71–86.
Demesure B, Sodzi N, Petit RJ (1995). A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Mol Ecol 4: 129–131.
Ditchfield AD (2000). The comparative phylogeography of Neotropical mammals: patterns of intraspecific mitochondrial DNA variation among bats contrasted to nonvolant small mammals. Mol Ecol 9: 1307–1318.
Dupanloup I, Schneider S, Excoffier L (2002). A simulated annealing approach to define the genetic structure of populations. Mol Ecol 11: 2571–2581.
Engel VL, Martins FR (2005). Reproductive phenology of Atlantic forest tree species in Brazil: an eleven year study. Trop Ecol 46: 1–16.
Ewing B, Green P (1998). Basecalling of automated sequencer traces using Phred II: error probabilities. Genome Res 8: 186–194.
Ewing B, Hillier L, Wendi M, Green P (1998). Basecalling of automated sequencer traces using Phred I: accuracy assessment. Genome Res 8: 175–185.
Excoffier L, Laval G, Schneider S (2005). Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinform Online 1: 47–50.
Fu YX (1997). Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147: 915–925.
Fu YX, Li WH (1993). Statistical tests of neutrality of mutations. Genetics 133: 693–709.
Gibbs P, Sassaki R (1998). Reproductive Biology of Dalbergia miscolobium Benth. (Leguminosae–Papilionoideae) in SE Brazil: the effects of pistillate sorting on fruit-set. Ann Bot 81: 735–740.
Gordon D, Abajian C, Green P (1998). Consed: A graphical tool for sequence finishing. Genome Res 8: 195–202.
Goudet J (2002). Fstat Version 2.9.3.2 Institute of Ecology, UNIL: Lausanne.
Graur D, Li WH (1999). Fundamentals of Molecular Evolution. Sinauer & Associates: Sunderland, Massachusetts.
Grivet D, Heinze B, Vendramin GG, Petit RJ (2001). Genome walking with consensus primers: application to the large single copy region of chloroplast DNA. Mol Ecol Notes 1: 345–349.
Hamilton MB (1999). Four primer pairs for the amplification of chloroplast intergenic regions with intraspecific variation. Mol Ecol 8: 521–523.
Hey J, Nielsen R (2004). Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila psudoobscura and D. persimilis. Genetics 167: 747–760.
Hey J, Nielsen R (2007). Integration within the Felsenstein equation for improved Markov chain Monte Carlo methods in population genetics. Proc Natl Acad Sci USA 104: 2785–2790.
IUCN (2008). IUCN Red List of Threatened Species. (www.iucnredlist.org) Downloaded on 20 February 2009.
Kelchner SA (2000). The evolution of non-coding chloroplast DNA and its application in plant systematics. Ann Missouri Bot Gard 87: 482–498.
Kuhner MK (2006). Lamarc 2.0: maximum likelihood and Bayesian estimation of population parameters. Bioinformatics 22: 768–770.
Kumar S, Tamura K, Nei M (2004). MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5: 150–163.
Ledru MP, Salatino MLF, Ceccantini G, Salatino A, Pinheiro F, Pintaud JC (2007). Regional assessment of the impact of climatic change on the distribution of a tropical conifer in the lowlands of South America. Diversity Distrib 13: 761–771.
Lira CF, Cardoso SRS, Ferreira PCG, Cardoso MA, Provan H (2003). Long-term population isolation in the endangered tropical tree species Caesalpinia echinata Lam. revealed by chloroplast microsatellites. Mol Ecol 12: 3219–3225.
Manni F, Guérard E (2004). Barrier version 2.2. Manual of the User. Population genetics team, Museum of Mankind (Musée de I’Homme): Paris.
Manni F, Guérard E, Heyer E (2004). Geographic patterns of (genetic, morphologic, linguistic) variation: how barriers can be detected using Monmonier's algorithm. Hum Biol 76: 173–190.
Monmonier MS (1973). Maximum-difference barriers: an alternative numerical regionalization method. Geogr Anal 3: 245–261.
Moraes-Barros N de, Silva JAB, Miyaki CY, Morgante JS (2006). Comparative phylogeography of the Atlantic forest endemic sloth (Bradypus torquatus) and the widespread three-toed sloth (Bradypus variegates) (Bracypodidae, Xenarthra). Genetica 126: 189–198.
Morellato LPC, Haddad CFB (2000). Introduction: The Brazilian Atlantic Forest. Biotropica 32: 786–792.
Moritz C (1994). Defining ‘Evolutionarily Significant Units’ for conservation. Trends Ecol Evol 9: 373–375.
Myers N, Mittermeier RA, Mittermeier CG, Fonseca GAB, Kent J (2000). Biodiversity hotsposts for conservation priorities. Nature 403: 853–858.
Myers N (1993). Biodiversity and precautionary principle. AMBIO 22: 74–79.
Nei M, Kumar S (2000). Molecular Evolution and Phylogenetics. Oxford University Press: New York.
Oliveira-Filho AT, Fontes MAL (2000). Patterns of floristic differentiation among Atlantic Forests in southeastern Brazil and the influence of climate. Biotropica 32: 793–810.
Pellegrino KCM, Rodrigues MT, Waite AN, Morando M, Yassuda YY, Sites Jr JW (2005). Phylogeography and species limits in the Gymnodactylus darwinii complex (Gekkonidae, Squamata): genetic structure coincides with river systems in the Brazilian Atlantic Forest. Biol J Linn Soc 85: 13–26.
Petit RJ, Mousadik AE, Pons O (1998). Identifying populations for conservation on the basis of genetics markers. Conserv Biol 12: 844–855.
Pons O, Petit RJ (1996). Measuring and testing genetic differentiation with ordered and unordered alleles. Genetics 144: 1237–1245.
Qiu YX, Guan BC, Fu CX, Comes HP (2009). Did glacials and/or interglacials promote allopatric incipient speciation in East Asian temperate plants? Phylogeographic and coalescent analyses on refugial isolation and divergence in Dysosma versipellis. Mol Phylogenet Evol 51: 281–293.
Ramos ACS, Lemos-Filho JP, Lovato MB (2009). Phylogeographical structure of the neotropical forest tree Hymenaea courbaril (Leguminosae: Caesalpinioideae) and its relationship with the vicariant Hymenaea stigonocarpa from Cerrado. J Hered 100: 206–216.
Ribeiro AR, Ramos ACS, Lemos-Filho JP, Lovato MB (2005). Genetic variation in remnant populations of Dalbergia nigra (Papilionoideae), an endangered tree from the Brazilian Atlantic Forest. Ann Bot 95: 1171–1177.
Rogers AR, Harpending HC (1992). Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9: 552–569.
Rozas J, Sànchez-DelBarrio JC, Messequer X, Rozas R (2003). Dnasp, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497.
Salgueiro F, Felix D, Caldas JF, Margis-Pinheiro M, Margis R (2004). Even population differentiation for maternal and biparental gene markers in Eugenia uniflora, a widely distributed species from the Brazilian coastal Atlantic rain forest. Divers Distrib 10: 201–210.
Smouse PE, Long JC, Sokal RR (1986). Multiple regression and correlation extension of the Mantel test of matrix correspondence. Syst Zoo 35: 627–632.
Taberlet P, Gielly L, Pautou G, Bouvet J (1991). Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol 17: 1105–1109.
Tajima F (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595.
Tchaicka L, Eizirik E, De Oliveira TG, Candido Jr JF, Freitas TRO (2007). Phylogeography and population history of the crab-eating fox (Cerdocyon thous). Mol Ecol 16: 819–838.
Thompson JD, Higgins DG, Gibson TJ (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680.
Weising K, Gardner RC (1999). A set of conserved PCR primers for the analysis of simple sequence repeat polymorphisms in chloroplast genomes of dicotyledonous angiosperms. Genome 42: 9–19.
Young A, Boyle T, Brown T (1996). The population genetic consequences of habitat fragmentation for plants. Trends Ecol Evol 11: 413–418.
Acknowledgements
We thank the three anonymous referees who made significant suggestions to improve this paper. RA Ribeiro received a doctoral fellowship and JP Lemos-Filho and MB Lovato received researcher fellowships from CNPq (Conselho Nacional de Desenvolvimento Tecnológico, Brazil). We also thank IBAMA (Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais Renováveis) and IEF-MG (Instituto Estadual de Florestas de Minas Gerais) for providing licenses for the collection of samples. We thank Renato M de Jesus (Vale do Rio Doce Reserve, Linhares, Espírito Santo), André Amorim (CEPLAC, Ilhéus, Bahia), Rosângela L Brandão, Renan ML Novaes, Júnia M Lousada and several other individuals, for assistance with the field work. We also thank Aline P Lorenz-Lemke and Gustavo S Cabanne for assistance with the data analyses. This study was supported by CNPq (Conselho Nacional de Desenvolvimento Tecnológico, Brazil) and FAPEMIG (Fundação de Amparo à Pesquisa do Estado de Minas Gerais, Brazil).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Ribeiro, R., Lemos-Filho, J., Ramos, A. et al. Phylogeography of the endangered rosewood Dalbergia nigra (Fabaceae): insights into the evolutionary history and conservation of the Brazilian Atlantic Forest. Heredity 106, 46–57 (2011). https://doi.org/10.1038/hdy.2010.64
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2010.64
Keywords
This article is cited by
-
Molecular markers applied to the genetic characterization of Dalbergia nigra: implications for conservation and management
Trees (2022)
-
What is the role of topographic heterogeneity and climate on the distribution and conservation of vascular epiphytes in the Brazilian Atlantic Forest?
Biodiversity and Conservation (2021)
-
From micro- to macroevolution: insights from a Neotropical bromeliad with high population genetic structure adapted to rock outcrops
Heredity (2020)
-
Historical climate changes and hybridization shaped the evolution of Atlantic Forest spinetails (Aves: Furnariidae)
Heredity (2019)
-
Late Pleistocene climate change shapes population divergence of an Atlantic Forest passerine: a model-based phylogeographic hypothesis test
Journal of Ornithology (2019)