Abstract
Parthenogenesis-inducing (PI) Wolbachia belong to a class of intracellular symbionts that distort the offspring sex ratio of their hosts toward a female bias. In many PI Wolbachia-infected species sex ratio distortion has reached its ultimate expression-fixation of infection and all-female populations. This is only possible with thelytokous PI symbionts as they provide an alternative form of reproduction and remove the requirement for males and sexual reproduction. Many populations fixed for PI Wolbachia infection have lost the ability to reproduce sexually, even when cured of the infection. We examine one such population in the species Trichogramma pretiosum. Through a series of backcrossing experiments with an uninfected Trichogramma pretiosum population we were able to show that the genetic basis for the loss of female sexual function could be explained by a dominant nuclear effect. Male sexual function had not been completely lost, though some deterioration of male sexual function was also evident when males from the infected population (created through antibiotic curing of infected females) were mated to uninfected females. We discuss the dynamics of sex ratio selection in PI Wolbachia-infected populations and the evolution of non-fertilizing mutations.
Similar content being viewed by others
Introduction
Symbiosis is a continuum of interaction incorporating all points between parasitism and mutualism. Intracellular symbionts in the bacterial genus Wolbachia span this continuum in the multitude of invertebrate hosts they infect. Wolbachia are maternally inherited reproductive symbionts from the alpha proteobacteria clade and as reproductive parasites are known to induce reproductive incompatibility, male killing, feminization, and thelytokous parthenogenesis in many arthropod hosts (Rousset et al., 1992; Stouthamer et al., 1993; Turelli, 1994; Hurst et al., 1999). Parasitic Wolbachia are generally considered facultative symbionts; meaning host organisms can survive and reproduce when cured. Wolbachia function in a more mutualistic manner in nematodes from the family Onchocercidae where experiments have shown they are essential for growth, longevity, and oogenesis (Bandi et al., 1999; Hoerauf et al., 1999). A symbiosis in the parasitoid Asobara tabida has been described as an obligatory dependence (Dedeine et al., 2001; Pannebakker et al., 2007), where an essential host reproductive function has been co-opted by Wolbachia. Hosts fail to develop or reproduce when cured of mutualistic or obligatory-dependent Wolbachia infections. Wolbachia in these species are obligate symbionts providing necessary developmental and reproductive functions.
The form of Wolbachia that induces thelytokous parthenogenesis (parthenogenesis-inducing (PI) Wolbachia), rather than providing an essential reproductive function, offers an alternative reproductive option: thelytokous parthenogenesis. The PI Wolbachia symbiosis is the only described Wolbachia phenotype that alters the mode of reproduction. Like other sex ratio distorting Wolbachia phenotypes, PI Wolbachia in haplodiploid organisms has been considered a facultative symbiont, unnecessary and often detrimental to host fitness (Stouthamer et al., 1990; Stouthamer and Luck., 1993). However, crossing experiments with several PI Wolbachia-infected species have shown Wolbachia infection in some species is required for female offspring production, making Wolbachia an obligate reproductive symbiont (Pijls et al., 1996; Jeong and Stouthamer, 2005; Kremer et al., 2009). Here, we describe the genetics underlying a proposed transition from facultative to obligate symbiont status as a consequence of PI Wolbachia's ability to alter the mode of reproduction.
By inducing thelytokous parthenogenesis in Wolbachia-infected females, female offspring production is no longer dependent on mating and egg fertilization. Trichogrammatid wasps, like other haplodiploid organisms, normally produce females by fertilizing eggs; males are derived directly from unfertilized eggs and are haploid. PI Wolbachia convert unfertilized haploid eggs into diploid embryos through an aborted first mitotic division, a process called gamete duplication (Stouthamer and Kazmer, 1994), and the resulting homozygous individual develops as an infected functional female. Many hymenopteran populations are fixed for PI Wolbachia and considered completely thelytokous (females produce only female offspring). Within and among Trichogrammatid wasp species PI Wolbachia infection frequencies range from low to complete fixation (Huigens and Stouthamer, 2003).
The complete fixation of PI Wolbachia infection in a population is often accompanied by the loss of female sexual function. Several studies have reported that mated females from PI Wolbachia-fixed populations fail to fertilize eggs with sperm from fully functional males (Zchori-Fein et al., 1995; Pijls et al., 1996; Arakaki et al., 2001; Jeong and Stouthamer, 2005; Pannebakker et al., 2005; Kremer et al., 2009). In mixed populations with low to intermediate infection frequencies infected females readily fertilize eggs when mated to conspecific males (Stouthamer et al., 1990). The reported data suggest that the population infection frequency selects for the loss of female sexual function.
There are three hypotheses explaining the observed loss of fertilization in PI Wolbachia-fixed populations and species. Once a population has reached a high level of PI Wolbachia infection the primary mode of reproduction is parthenogenesis and alleles coding for sexual traits are expected to accumulate mutations at a neutral rate (Carson et al., 1982). Both male and female sexual functions are subject to deterioration by genetic drift according to the mutation accumulation hypothesis. The costly female trait hypothesis (Pijls et al., 1996) suggests that once PI Wolbachia has spread parthenogenesis through a population, obsolete female sexual traits that are physiologically costly to maintain will be selected against (the loss of costly female sexual traits will result in increased fitness); male sexual traits will deteriorate at a neutral rate through mutation accumulation. The virginity mutation hypothesis (Huigens and Stouthamer, 2003) proposes sex ratio selection (Fisher, 1930; Hamilton, 1967) is responsible for the loss of female sexual function, regardless of physiological cost, and the maintenance of male sexual function as PI Wolbachia spreads through populations.
When PI Wolbachia enters a population it introduces a deviation from ‘optimal’ sex ratios (Fisher, 1930; Hamilton, 1967). As the infection spreads and the population sex ratio becomes more female biased the value of outcrossing males increases. Females with reduced fertilization frequency in haplodiploid populations gain the sex ratio benefit of more valuable sons. A female's fitness in such populations is inversely related to fertilization frequency; lower fertilization frequency genotypes outcompete higher fertilization frequency genotypes (Huigens and Stouthamer, 2003).
The virginity mutation hypothesis predicts and unpublished simulation models have confirmed that alleles decreasing fertilization rates (‘virginity’ alleles) are positively selected in PI Wolbachia-infected populations and sweep to fixation. Once a population is fixed for the ‘virginity’ allele Wolbachia-induced thelytokous parthenogenesis becomes the sole means of female production. Therefore, the Wolbachia infection spreads to all females and the population becomes functionally parthenogenetic. In a population fixed for Wolbachia whose sole means of reproduction is parthenogenesis, alleles coding for male sexual traits would be subject to mutation accumulation; and superfluous female sexual traits will be eliminated if they are costly to maintain (Pijls et al., 1996). Therefore, both costly female trait and virginity mutation hypotheses are not necessarily mutually exclusive.
It is assumed in the virginity mutation hypothesis that, in normal sexual populations mutations are present at low frequencies that cause females expressing these mutations to fertilize a lower proportion of their eggs. In an uninfected population such mutations may not be selected against, depending on the mating structure of the population. In randomly mating populations, selection against these mutations will not be very strong. However, in Trichogramma with its high frequencies of sibmating, we expect strong selection against fertilization rate mutations. Once the infection frequency in a population has reached a substantial level, mutations that reduce the fertilization proportion will be selected and the lower the fertilization rate induced by the mutation the larger the selective advantage. Small influence ‘virginity’ mutations may act in a quantitative manner, however mutations with major effects on the fertilization rate will spread and bring Wolbachia and the mutant allele to fixation faster.
Here, we determine whether in a population of Trichogramma pretiosum fixed for the PI-Wolbachia infection, females express the non-fertilization phenotype and the genetic basis for this trait. It is important to note here that the use of the word virginity strictly refers to the loss of female sexual function. Sexual function involves many physiological and behavioral traits, of which many may have a polygenic basis (Carson et al., 1982). However, only one of these traits need be affected by a mutant allele to induce the non-fertilization phenotype. Using a crossing protocol we show the genetic basis for such an allele in a population of Wolbachia-infected Trichogramma pretiosum.
Materials and methods
Insect cultures
Trichogramma pretiosum stocks
Trichogramma pretiosum-infected and -uninfected cultures originated in Peru and California, respectively. Infected Trichogramma pretiosum cultures were established from collections at high altitude locations in Peru where the Trichogramma pretiosum population is fixed for Wolbachia infection. Uninfected Trichogramma pretiosum from California were collected from Heliothus virescens eggs on experimental tomato plants at the University of California South Coast Research and Extension Center in Irvine, California in 2004. All Trichogramma pretiosum cultures originated from single females (mated if uninfected) and maintained in the laboratory on Ephestia kuehniella eggs (Beneficial Insectary, Redding, CA, USA) at 24 °C, L:D=16:8 and 50% relative humidity.
Species identification
Trichogramma pretiosum species were identified using protocols described in Stouthamer et al. (1999). Sequencing of the ITS2 gene region was conducted at the University of California Riverside Genomics Institute Core Instrumentation Facility using the Applied Biosystems 3730 DNA analyzer with Big-Dye version 3.1 kit (Applied Biosystems, Foster City, CA, USA). Sequences were aligned using BioEdit version 6.0.7 (Hall, 1999) and the identification of the species was confirmed by comparing the DNA sequences to already known ITS2 sequences for this species (Stouthamer et al., 1999).
Wolbachia infection status
The infection status of the experimental lines was tested by virgin reproduction and molecular methods. PI Wolbachia infection can be detected by the female-biased offspring sex ratio of virgin females. Virgin experimental lines originating from the Peruvian and Californian cultures produced 97 and 0% female offspring, respectively.
As further proof of infection status, PCR primers specific for the Wolbachia B strain were used according to previously described procedures (Werren et al., 1995). All females sampled from the Peruvian culture tested positive for Wolbachia infection. All females from the Californian culture tested negative for infection.
Fertilization frequency of infected and uninfected Trichogramma pretiosum
As haploid males develop directly from unfertilized eggs and females from fertilized eggs, the fertilization frequency of haplodiploid organisms can be approximated by the observed offspring sex ratio. Therefore, our offspring sex ratio values were determined by calculating the proportion of female offspring (female offspring/total offspring) to reflect female fertilization frequencies. To determine the fertilization frequency of the females of the infected line, we tried to cure this line of their infection. This was done by subjecting 20 isofemale lines from the Wolbachia infected culture to the antibiotic rifampicin, which is known to eliminate Wolbachia (Stouthamer et al., 1990). Newly emerged infected females were fed a 0.5% g ml−1 antibiotic/honey solution for 1 day. Each day new host eggs were supplied and the previous days host eggs were incubated. All females had access to antibiotic honey solution throughout the 8 days of oviposition. Next generation offspring emerging from the daily hosts were observed for sex ratio and those broods that showed a sex ratio approaching 50% were again given the antibiotic treatment described above. This curing procedure lasted no more than three generations before some lines expired due to an absence of female offspring. By generation 7 all 20 lines had died out. Despite numerous attempts to adjust the antibiotic concentration no sexually reproducing lines from the infected culture established.
Trichogramma wasps infected with PI Wolbachia will often fertilize their eggs at frequencies similar to uninfected females (Stouthamer and Kazmer, 1994). The fertilization rate of eggs can be determined genetically by determining the fraction of female offspring that carry a nuclear marker from the father. Two microsatellite markers specifically designed for Trichogramma species (TAC 47: forward CTACGGCGACAATTGCCAC, reverse CATCTTGGTCGAACCGAGCAG; TTG 46: forward GATGTTTACTTCGCAGGCCGC, reverse CTACGGGGCATACGATATGTG; Huigens, 2003) were used to test paternally inherited alleles in the female offspring from mated infected mothers using the following thermocycle conditions: 94 °C for 3 min, then 35 cycles of 94 °C for 45 s, 50 °C for 45 s, 72 °C for 45 s, and a final extension at 72 °C for 3 min. Fourteen virgin infected females were isolated and given virgin males from the uninfected population. After mating was observed each female was allowed to oviposit. One day after mating, all males were removed and after 6 days of oviposition the females were removed. Both males and females were preserved in 95% EtOH. Between 5 and 8 female offspring from 13 of the 14 infected mated females (one female died without reproducing) were genotyped using the microsatellite markers. To determine the fertilization frequency of the uninfected California culture 65 virgin females were mated to virgin males from the same culture and allowed to oviposit on E. kuehniella hosts for 6 days. The number and sex of the offspring were determined after their emergence.
Crossing experiments
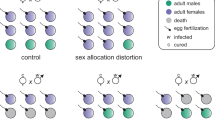
A series of crosses aimed at determining the genetic basis of non-fertilization are described in Figure 1. The nomenclature used was meant to reflect the nuclear genetic background and ploidy of the crosses. For instance, females from the California culture are characterized as ‘CC’, males from the Peruvian culture as ‘P’; hybrid females resulting from a cross between California females and Peru males as ‘CP’, and recombinant males produced by CP females as ‘c/p’. Introgression of the nuclear genome from the infected line into the uninfected line was accomplished by mating males from the infected line with females of the uninfected line. Males from the infected line were produced by feeding infected females antibiotics, thus curing eggs of Wolbachia and resulting in mostly male offspring. Males and females were isolated in 12 mm × 75 mm glass culture tubes for 24 h, after which males were removed and females were given E. kuehniella host eggs ad libitum for 6 days.
(a) Hybrid females (CP) resulting from the cross between California (CC) females and Peru (P) males are expected to produce male-biased offspring sex ratios (female offspring/total offspring) according to the dominant model of inheritance for the non-fertilization allele, and female-biased sex ratios according to the recessive model of inheritance. (b) Assuming a dominant mode of inheritance, one-locus and two-loci models of inheritance can be distinguished by the modal distribution of offspring sex ratios in the D1 cross (a cross between control California males (C) and daughters of California females (CC) and recombinant males (c/p) cross. The one-locus model results in a bimodal distribution and the two-loci model results in a trimodal distribution assuming strictly additive effects of the non-fertilization alleles.
The logic of the experiments was to (1) introgress the Peruvian (non-fertilization) genome into the California genome, and (2) determine the egg fertilization rate of the introgressed females. The potential methods of inheritance for the non-fertilization trait in the Trichogramma pretiosum line from Peru can be broken down into nuclear/cytoplasmic, dominant/recessive, and one-locus/multi-loci models. The cytoplasmic model assumes that cytoplasmically inherited Wolbachia directly interacts with the fertilization process. Two lines of evidence suggest that this is not the case in species infected with PI Wolbachia: (1) In populations where the PI Wolbachia infection has progressed to fixation, females cured of Wolbachia infection with antibiotics still fail to fertilize the eggs (Arakaki et al., 2001; Jeong and Stouthamer, 2005; Pannebakker et al., 2005; Pijls et al., 1996; Kremer et al., 2009), and (2) in populations where the PI Wolbachia infection has not reached fixation infected females continue to fertilize eggs (Stouthamer et al., 1990). Evidence supporting the nuclear model is found in Jeong and Stouthamer (2005) for Telonomus nawai, where nuclear introgression induced the non-fertilization phenotype in an uninfected population. Assuming a nuclear mode of inheritance for the trait in Trichogramma pretiosum we can distinguish between dominant and recessive models through a crossing experiment between females from the California uninfected population (CC) and males derived from the Peruvian-infected population (P) (Figure 1a). If the trait is inherited in a dominant manner we expect that hybrid females (CP), derived from the CC × P cross, will not fertilize their eggs and will produce male-biased offspring sex ratios. If the trait is inherited in a recessive manner we expect hybrid females to fertilize their eggs at a normal (control) frequency and produce wild-type offspring sex ratios.
If the non-fertilization trait is inherited in a dominant manner, the question of number of dominant alleles with major effect arises. This is tested by using recombinant haploid males (c/p) that are mated with California females (CC). If there is only a single locus involved in the trait then recombinant males are either carriers of the non-fertilization trait or have the wild-type allele. Consequently, the females fathered by recombinant males (Cc/p) should either express the non-fertilization trait and fertilize their eggs at a low frequency or have normal egg fertilization rates; thus producing a bimodal distribution of offspring sex ratios among the daughters of wild-type California females (CC) mated with recombinant males (c/p). We refer to the cross of such daughters with control males (C) as the Dominance 1 (D1) cross (Figure 1b).
The alternative model involving more loci is more complex, but if we assume a simple two-loci model where each locus contributes equally to the low fertilization rate, then we would expect a female that carries a non-fertilization allele at one of the two loci to fertilize her eggs at half the rate of a wild-type female. Under this model, females fathered by recombinant males would fall into four types: females homozygous for the wild-type alleles at both loci, females homozygous for the wild type at one of the two loci and heterozygous at the other locus, and finally females heterozygous at both loci. Assuming unlinked loci, 25% of the females would be expressing the wild-type phenotype, 50% would express the phenotype where the fertilization rate is half of the wild-type fertilization rate, and 25% of the females would express the non-fertilization phenotype. This should result in a trimodal distribution of offspring sex ratios. The difference between the one and two-loci models is the bimodal distribution expected under the single-locus model for the offspring produced by the females produced in the cross between a CC female and a recombinant male, whereas under the two-loci model a trimodal distribution is expected (Figure 1b).
The expected frequency distributions shown in Figure 1 are intended to illustrate the expected outcome of different inheritance models, dominant versus recessive and single-locus versus two-loci models. These distributions are meant to give a general indication of offspring sex ratio frequencies given either a one-locus or two-loci dominant model for the non-fertilization trait. There are no a priori exact frequency distribution expectations other than a female-biased offspring sex ratio for control females, as mentioned in previous analyses for Trichogramma pretiosum (Kazmer, 1992; Stouthamer and Kazmer, 1994) and a male-biased sex ratio distribution for hybrid females carrying a dominant non-fertilization allele.
D experiments
The choice of a dominant model was governed by the observed results in the initial cross between F1 hybrid females and males from both populations (Figure 2b; Table 1). For the D experiments we were able to generate expected distributions based on the results of the crosses involving control and hybrid females given an assumed one-locus dominant model that could be analyzed quantitatively. To test the dominance of the non-fertilization trait the offspring of the CC × c/p cross (California, uninfected sexual females mated to recombinant males) were crossed and tested for fertilization frequency. This test was conducted twice (D1 experiment, n=77, 137) with the results of the second test (n=137) used for further analysis. In this cross the resultant females (Cc/p) were effectively 1/2 hybrids because of the recombined paternal genome. If functional virginity is controlled by a single dominant locus that segregates in Mendelian manner then the offspring sex ratios of the cross involving Cc/p females mated to C males should show a bimodal distribution corresponding to the control (CC × C) and hybrid female (CP × C) offspring sex ratio distributions taken together (Figures 2a and b). Expected distributions for the D1 and D2 male-biased crosses were derived by multiplying the observed offspring sex ratio class frequencies by 0.5 for both the CC × C and CP × C crosses, then adding the offspring sex ratio class frequencies from both crosses. The expected distribution for the D2 female-biased cross was taken directly from the observed distribution for the control CC × C cross.
(a, b) Observed distributions of the control (CC × C) and hybrid female (CP × C) crosses are shown. The crosses are designated by the diploid genotype females (CC and CP) and haploid genotypes of males (C). The x axis represents the offspring sex ratio (female offspring/total offspring) classes such that the 0.0 class represents all offspring sex ratio frequency values lower than 0.1, the 0.1 class represents all offspring sex ratio frequency values greater than or equal to 0.1 and lower than 0.2.
The offspring of the second D1 experiment were used for validation of a single-locus dominance effect on fertilization (D2 female/male bias; Supplementary Table S1 in Supplementary Material; Figures 3a–f). If the dominant mutant allele for loss of fertilization segregates in Mendelian manner, the allele should be present in half of the female offspring of the females expressing the trait ((CP) genotype females), that is mothers that produce offspring sex ratios in the 0–0.39 female proportion range; the left side of the bimodal distribution, assuming incomplete dominance. Therefore, female offspring from this side of the bimodal distribution should replicate the bimodal offspring sex ratio distribution (Figure 3c). Female offspring from mothers that do not express the loss of fertilization trait (CC genotypes) should produce offspring sex ratios no different than control females (Figure 3d). A complete list of all experimental crosses is presented in Supplementary Table S2.
(a–f) Expected and observed distributions of the D1 and D2 crosses assuming a dominant non-fertilization effect. The expected distributions assume a high level of dominance for the non-fertilization allele. Expected distributions for the D1 and D2 male-biased crosses were derived by multiplying the observed offspring sex ratio class frequencies by 0.5 for both the CC × C and CP × C crosses, then adding the offspring sex ratio class frequencies from both crosses. The expected distribution for the D2 female-biased cross was taken directly from the observed distribution for the control CC × C cross. The x axes represent offspring sex ratio classes calculated as female offspring/total offspring and grouped by frequency increments of 0.1.
Degree of dominance
Using methods described in Lynch and Walsh (1998) hypotheses regarding the degree of dominance for the non-fertilization trait can be illustrated by dominance coefficient values for the California control (sexual) × Peru parthenogenetic hybrid female (CP) fertilization phenotype. Dominance values in the −1 to 0 range represent the impact of the parthenogenetic Peruvian genome, with a −1 value representing complete dominance. Dominance values in the 0–1 range represent the impact of the control Californian genome, with a value of 1 representing complete dominance.
Statistics
Offspring sex ratio, male, female, and total offspring production, and pupal mortality were analyzed using ANOVA, Fisher's Protected Least Significant Difference, and Scheffe's post hoc comparison tests. Offspring sex ratio distributions were subjected to Kolmogorov–Smirnov goodness of fit and G tests. Analyses were conducted using StatView (SAS Institute Inc., Cary, NC, USA) and R programming language (Ihata and Gentleman, 1996).
Results
Fertilization frequency of experimental cultures
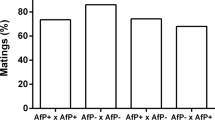
The offspring sex ratio (proportion females=females/total offspring) of the uninfected Californian (C) culture was used as a measure of fertilization frequency. Matings between CC females and C males (CC × C, n=65) produced a mean offspring sex ratio of 0.675±0.11s.d. (67.5% female offspring). The offspring sex ratio distribution for the C culture shows that over 70% of all females produce offspring sex ratios in the 0.6 and 0.7 classes (Figure 2a). The lowest sex ratio class was 0.3 (30–39% female offspring); the highest was 0.9 (90–99% female offspring).
The offspring sex ratio of the infected Peruvian (P) culture could not be used to assess the fertilization frequency of PP females because of our inability to establish a cured culture, despite the presence of males in antibiotically treated cultures and observed mating. Therefore, microsatellites were used to determine whether infected PP females fertilized any eggs when mated to C males. A total of 94 female offspring representing between 5 and 8 offspring from 13 mated PP females showed no successful fertilization of eggs. Microsatellite analysis plus the inability to establish a cured culture, despite repeated attempts, showed that females from the Peruvian parthenogenetic population did not successfully fertilize any eggs.
Introgression
Total offspring production values for uninfected CC females mated to males from the Peruvian (P) and California (C) populations were 38.5±14.1 and 44.7±12.3, respectively (Table 1). Mean fertilization frequency for CC females mated to P males (CC × P cross) as measured by the offspring sex ratio was 0.50±0.19 (50% female offspring). The variance of the CC × P cross offspring sex ratios ( =0.038) was more than twice the variance found in the control CC × C cross (
=0.038) was more than twice the variance found in the control CC × C cross ( =0.012).
=0.012).
The hybrid females (CP), resulting from the CC × P cross, backcrossed to males from both parental backgrounds produced offspring sex ratios considerably lower than control females (CC) (Table 1). Hybrid females mated to control males (CP × C cross) produce male-biased offspring sex ratios (Figure 2b).
Among the various matings mean offspring sex ratios varied in the following order: (CC × C, CC × c/p, D2 female)>(CC × P)>(D1, D2 male)>(CP × C)> (CP × P) (Table 1).
Offspring production values were higher for the D-test crosses when compared with all other crosses, except CC × c/p (Table 1). Hybrid females mated to P males and PPV females produced fewer offspring than all other crosses. The reduction in average offspring between control CC females and hybrid (CP) females is a product of fewer females produced by the hybrid females. All hybrid females produced more male offspring than all control females except virgin CC females (Scheffe's P<0.0001). All hybrid females produced statistically similar numbers of female offspring.
D experiments
Roughly half of the offspring sex ratio distribution found in the test for dominance (D1 experiment, n=137) was absent in the control cross (Figures 2a and 3b). Offspring sex ratio values from the D1 cross in the 0–0.39 range (higher male production) accounted for 50.1% of the frequency distribution. For comparison, in the control cross the offspring sex ratios in this range only accounted for 1.5% of the frequency distribution (Figure 2a), and 83.8% of the observations among hybrid females in the CP × C cross (Figure 2b) were in this range. The offspring sex ratio distribution from the second D1 cross (N=137) was statistically no different from the first D1 cross (Kolmogorov–Smirnov, P=0.2956; Supplementary Table S3). Both D1 cross distributions were significantly different (individually and pooled, G-test, P<0.023) than the expected distribution (Figure 3a; Supplementary Table S3).
Sibling-mated females selected from the respective modes of the bimodal distribution seen in the second D1 experiment made up the D2 male bias (females selected from the 0–0.39 offspring sex ratio classes, n=61) and the D2 female bias (females selected from the 0.4–0.99 offspring sex ratio classes, n=27) crosses. The D2 male bias cross offspring sex ratio distribution (Figure 3e) is not statistically different from the first D1 experiment (Kolmogorov–Smirnov, P=0.8767; Supplementary Table S4), but showed a statistical difference from the second D1 experiment, from which it was taken, (Kolmogorov–Smirnov, P=0.0425; Supplementary Table S4) and the expected D1 distribution (G=19.653, 9 d.f., G-test, P<0.02; Supplementary Table S4). There is no statistical difference between the D2 female bias (Figure 3f) and the CC × C offspring sex ratio distributions (Kolmogorov–Smirnov, P=0.3159; Supplementary Table S4). There were no significant differences in offspring production among the D crosses (ANOVA, P=0.9171; Supplementary Table S5).
Degree of dominance
A purely additive effect of introgression of the genome from the parthenogenetic (infected) population on the control sexual (uninfected) genome would result in a midpoint fertilization percentage in the F1 hybrids of 33.75% (Figure 4). Instead, we found a fertilization percentage of 14.7% when hybrid females were mated to control males and 3.6% when mated to males antibiotically derived from parthenogenetic females. This represented a 78 and 93% reduction in fertilization when compared with control females. A dominance coefficient of −0.64 indicated a high degree of dominance for the parthenogenetic background in terms of the non-fertilization trait in Trichogramma pretiosum.
The genotypic values for fertilization percentage are shown for the experimental lines used (PP × C, Peru, parthenogenetic female crossed with a control male; CC × C, California, control female crossed with a control male; CP × C obs, observed hybrid female crossed with a control male; CP × C exp, expected hybrid female crossed with a control male). The actual values are PP × C=0, CC × C=67.5%, CP × C exp=33.75%, and CP × C obs=14.7%.
Discussion
Our investigation into the non-fertilization trait in Trichogramma pretiosum began with an assumption of genetic recessivity, similar to that found in Telonomus nawai (Jeong and Stouthamer, 2005), and a protocol that required at least two generations of backcrossing. The observed dominant effect on fertilization in first generation hybrid females was confirmed by the following D1 and D2 crosses. The results of the D experiments support the dominant single-locus model explaining the loss of female sexual function. Rather than the reproductive incompatibility observed in Trichogramma deion where female offspring simply died and no change occurred in male offspring production (Stouthamer et al., 1996), hybrid females in our study produce only male offspring with no significant change in total offspring production; a result that is concordant with the virginity mutation hypothesis—a significant reduction in fertilization. However, alternative explanations for the change in female fertilization behavior and offspring sex ratios could be selection for the loss of costly female traits in an infected parthenogenetic population (Pijls et al., 1996), or mutation accumulation in pathways associated with egg fertilization. Our results do not directly distinguish between the possible mechanisms.
The development and full functionality of recombinant males in the CP × C and CC × c/p crosses does not support hybrid breakdown because of incompatible genetic interactions. The offspring sex ratio range 0–0.39, which is absent in the control CC × C cross but present in the D1, D2 male bias cross, and hybrid female (CP) crosses, appears to result from a dominant effect of the P genome. The predicted bimodal offspring sex ratio distribution given a dominant genetic effect was observed (Figures 1, 2, 3), though the predicted and observed bimodal distributions were statistically different for the D-test crosses.
The observed reduced offspring production of the mated hybrid females together with reduced female production (Table 1) point to a possible interaction between the C and P genomes that have some deleterious effect on female development. However, this cannot completely explain the differences in offspring sex ratios and egg fertilization between control and hybrid females. Even when the female offspring, assumed lost through developmental mortality, are replaced there is still a large difference that results in a reversal of offspring sex allocation between control and hybrid females (67.5% female offspring for the control versus 31.9% female offspring for hybrid).
A reversal of offspring sex allocation when PI Wolbachia-infected and -uninfected populations are introgressed was shown for Telenomus nawai (Jeong and Stouthamer, 2005). However, the inheritance of the non-fertilization trait in Telonomus nawai showed a recessive pattern that could be explained as a single gene of major effect with modifiers or two linked loci. Our results for Trichogramma pretiosum also suggest a major effect gene associated with fertilization (functional virginity), but expressed in a dominant manner. The lack of statistical fit in the D1 and D2 male-biased dominance tests also may be a consequence of genetic modifiers. In highly inbred populations such as Trichogramma pretiosum (Kazmer, 1992) the difference in selection pressures for dominant and recessive low frequency fertilization alleles is significantly decreased. The effect of inbreeding results in levels of allelic homozygosity and phenotypic expression that differ little between recessive and dominantly expressed alleles (Hedrick and Parker, 1997). Therefore, the observed dominant ‘virginity’ effect of the Peruvian parthenogenetic genome is not unexpected.
The dominant effect on female sexual function we observe in the hybrid females of this study may indeed be a result of a selective sweep in the parthenogenetic population for the removal of costly female sexual traits (Pijls et al., 1996). Mutation accumulation in sexual traits in a parthenogenetic population would likely result in some loss of function for both male and female reproductive traits (Carson et al., 1982). However, the observed pattern in PI Wolbachia-infected populations suggest female-limited reproductive traits are more likely to decay before male reproductive function is lost (Zchori-Fein et al., 1995; Pijls et al., 1996; Arakaki et al., 2001; Jeong and Stouthamer, 2005; Pannebakker et al., 2005; Kremer et al., 2009). If the loss of female sexual function is a product of selection for mutations in costly female reproductive traits then we have to assume the genes that code for these traits are positively selected in a sexual context and negatively selected in a parthenogenetic context; that is, the traits carry a fitness advantage in a sexual population and a fitness cost in a parthenogenetic population. This could be considered a type of antagonistic pleiotropy associated with the mode of reproduction rather than reproduction itself (Hedrick, 1999). The question becomes which female sexual traits are intrinsically costly?
The virginity mutation hypothesis is based on a selective advantage for the loss of female sexual function traits regardless of associated physiological fitness benefits. Furthermore, the virginity mutation hypothesis, which is based on the fitness advantage of rare males in a population with a sweeping PI Wolbachia infection, requires the maintenance of male sexual function (Huigens and Stouthamer, 2003). Any gene allowing females in an infected population to produce more male offspring that can take advantage of surplus mating opportunities provided by unmated females will be selected accordingly. The two scenarios, selection for mutations in costly female sexual traits and selection for ‘virginity’ mutations, are not mutually exclusive. The difference between the two scenarios is a selective advantage for the loss of female sexual function and maintenance of male sexual function in the initial stages of Wolbachia infection according to the virginity mutation hypothesis, and the loss of female sexual function by selection and male sexual function by genetic drift according to the costly female trait hypothesis. The virginity mutation hypothesis gives an adaptive explanation for the maintenance of male sexual function. The costly female trait hypothesis explains the maintenance of male sexual function as a function of drift having less influence over evolutionary forces, resulting in selection for the loss of costly female traits preceding the loss of male sexual traits by drift. The loss of female sexual function in either context is not the product of neutral or indirect selection processes (Fong et al., 1995; Maughan et al., 2007), but the result of selection imposed by a bacterial infection that initially induces facultative thelytokous parthenogenesis; only later evolving into an obligate parthenogenetic symbiont.
In the populations studied where PI Wolbachia has swept to fixation it appears the sexual functionality of females and antibiotic-induced males conform to the predictions of the virginity mutation hypothesis (Pijls et al., 1996; Arakaki et al., 2000; Weeks and Breeuwer, 2001; Jeong and Stouthamer, 2005; Pannebakker et al., 2005; Kremer et al., 2009), namely, that females are sexually non-functional and males are sexually functional. Once the Wolbachia infection and virginity mutation are fixed male sexual function is subject to drift, mutation accumulation, and ultimate deterioration (Muller, 1932; Carson et al., 1982). A high degree of male sexual function variance in a PI Wolbachia-fixed population of Leptopilina clavipes was responsible for the variation seen in offspring sex ratios (Pannebakker et al., 2005). Similar results are reported here with sex ratio variance doubling in crosses between control females (CC) and Peruvian males derived from the parthenogenetic line (P). In Muscidifurax uniraptor, a species completely fixed for PI Wolbachia, male sexual function is completely lost and female sexual function has been eliminated both behaviorally and morphologically—suggesting a long history of PI Wolbachia infection (Gottlieb and Zchori-Fein, 2001). Our results also suggest male sexual function, though not completely lost, has deteriorated (Table 1, compare CC × C with CC × P, and CP × C with CP × P). Alternatively, deleterious genomic interactions may be responsible for the observed results. If male sexual function variance in Trichogramma pretiosum is a product of incompatible genomic interactions we might expect to see some deleterious sexual function in the recombinant males. However, this is not the case. The offspring and offspring sex ratio values for recombinant males mated to control females were statistically similar to control males (Table 1).
Similarities between the A. tabida and Trichogramma pretiosum symbioses are informative. Both systems have evolved reproductive dependence on Wolbachia. However, the two trajectories are distinct. In A. tabida, Wolbachia takes over a pre-existing host function oogenesis (Dedeine et al., 2001; Pannebakker et al., 2007; Vavre et al., 2008). In Trichogramma pretiosum Wolbachia provides a novel function, thelytokous parthenogenesis through gamete duplication (Stouthamer and Kazmer, 1994; Stouthamer et al., 1999). Both scenarios lead to the obligate dependence on Wolbachia for reproductive function. These similarities and differences highlight the dexterity of Wolbachia when it comes to achieving obligate symbiont status.
References
Arakaki N, Miyoshi T, Noda H (2001). Wolbachia-mediated parthenogenesis in the predatory thrips Franklinothrips vespiformis (Thysanoptera: Insecta). Proc R Soc Lond B Biol Sci 268: 1011–1016.
Arakaki N, Noda H, Yamagishi K (2000). Wolbachia-induced parthenogenesis in the egg parasitoid Telenomus nawai. Entomol Exp Appl 96: 177–184.
Bandi C, McCall JW, Genchi C, Corona S, Venco L, Sacchi L (1999). Effects of tetracycline on the filarial worms Brugia pahangi and Dirofilaria immitis and their bacterial endosymbionts Wolbachia. Int J Parasitol 29: 357–364.
Carson HL, Chang LS, Lyttle TW (1982). Decay of female sexual behavior under parthenogenesis. Science 218: 68–70.
Dedeine F, Vavre F, Fleury F, Loppin B, Hochberg ME, Boulétreau M (2001). Removing symbiotic Wolbachia bacteria specifically inhibits oogenesis in a parasitic wasp. Proc Natl Acad Sci USA 98: 6247–6252.
Fisher RA (1930). The Genetical Theory of Natural Selection. Oxford University Press: Oxford.
Fong DW, Kane TC, Culver DC (1995). Vestigialization and loss of nonfunctional characters. Annu Rev Ecol Syst 26: 249–268.
Gottlieb Y, Zchori-Fein E (2001). Irreversible thelytokous reproduction in Muscidifurax uniraptor. Entomol Exp Appl 100: 271–278.
Hall TA (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98.
Hamilton WD (1967). Extraordinary sex ratios. Science 156: 477–488.
Hedrick PW (1999). Antagonistic pleiotropy and genetic polymorphism: a perspective. Heredity 82: 126–133.
Hedrick PW, Parker JD (1997). Evolutionary genetics and genetic variation of haplodiploids and X-linked genes. Annu Rev Ecol Syst 28: 55–83.
Hoerauf A, Nissen-Paehle K, Schmetz C, Henkle-Duehrsen K, Blaxter ML, Buettner DW et al. (1999). Tetracycline therapy targets intracellular bacteria in the filarial nematode Litomosoides sigmodontis and results in filarial infertility. J Clin Invest 103: 11–18.
Huigens ME (2003). On the evolution of Wolbachia-induced parthenogenesis in Trichogramma wasps. PhD dissertation, Wageningen University, the Netherlands.
Huigens ME, Stouthamer R (2003). Parthenogenesis associated with Wolbachia. In: Bourtzis K, Miller TA (eds). Insect Symbiosis. CRC Press: Boca Raton, pp 247–266.
Hurst GDD, Jiggins FM, von der Schulenburg JHG, Bertrand D, West SA, Goriacheva II et al. (1999). Male-killing Wolbachia in two species of insect. Proc R Soc Lond B Biol Sci 266: 735–740.
Ihata R, Gentleman R (1996). R: a language for data analysis and graphics. J Comput Graph Stat 5: 299–314.
Jeong G, Stouthamer R (2005). Genetics of female functional virginity in the Parthenogenesis-Wolbachia infected parasitoid wasp Telenomus nawai (Hymenoptera:Scelionidae). Heredity 94: 402–407.
Kremer N, Charif D, Henri H, Bataille M, Prevost G, Kraaijeveld K et al. (2009). A new case of Wolbachia dependence in the genus Asobara: evidence for parthenogenesis induction in Asobara japonica. Heredity 103: 248–256.
Kazmer DJ (1992). The evolution of reproductive strategies in Trichogramma and its relationship to biological control. PhD dissertation, University of California, Riverside.
Lynch M, Walsh B (1998). Genetics and Analysis of Quantitative Traits. Sinauer Associates Inc.: Sunderland, MA, pp 69–71.
Maughan H, Masel J, Birky CW, Nicholson WL (2007). The roles of mutation accumulation and selection in loss of sporulation in experimental populations of Bacillus subtilis. Genetics 177: 937–948.
Muller HJ (1932). Some genetic aspects of sex. Am Nat 66: 118–138.
Pannebakker BA, Loppin B, Elemans CPH, Humblot L, Vavre F (2007). Parasitic inhibition of cell death facilitates symbiosis. Proc Natl Acad Sci USA 104: 213–215.
Pannebakker BA, Schidlo NS, Boskamp GJF, Dekker L, Van Dooren TJM, Beukeboom LW et al. (2005). Sexual functionality of Leptopilina clavipes (Hymenoptera:Figitidae) after reversing Wolbachia-induced parthenogenesis. J Evol Biol 18: 1019–1028.
Pijls JWAM, Van Steenbergen HJ, Van Alphen JJM (1996). Asexuality cured: the relations and differences between sexual and asexual Apoanagyrus diversicornis. Heredity 76: 506–513.
Rousset F, Bouchon D, Pintureau B, Juchault P, Solignac M (1992). Wolbachia endosymbionts responsible for various alterations of sexuality in arthropods. Proc R Soc Lond B Biol Sci 250: 91–98.
Stouthamer R, Breeuwer JAJ, Luck RF, Werren JH (1993). Molecular-identification of microorganisms associated with parthenogenesis. Nature 361: 66–68.
Stouthamer R, Hu JG, van Kan FJPM, Platner GR, Pinto JD (1999). The utility of internally transcribed spacer 2 DNA sequences of the nuclear ribosomal gene for distinguishing sibling species of Trichogramma. Biocontrol 43: 421–440.
Stouthamer R, Kazmer DJ (1994). Cytogenetics of microbe-associated parthenogenesis and its consequences for gene flow in Trichogramma wasps. Heredity 73: 317–327.
Stouthamer R, Luck RF (1993). Influence of microbe-associated parthenogenesis on the fecundity of Trichogramma deion and T. pretiosum. Entomol Exp Appl 67: 183–192.
Stouthamer R, Luck RF, Hamilton WD (1990). Antibiotics cause parthenogenetic Trichogramma (Hymenoptera, Trichogrammatidae) to revert to sex. Proc Natl Acad Sci USA 87: 2424–2427.
Stouthamer R, Luck RF, Pinto JD, Platner GR, Stephens B (1996). Non-reciprocal cross-incompatibility in Trichogramma deion. Entomol Exp Appl 80: 481–489.
Turelli M (1994). Evolution of incompatibility-inducing microbes and their hosts. Evolution 48: 1500–1513.
Vavre F, Kremer N, Pannebakker BA, Loppin B, Mavingui P (2008). Is symbiosis evolution influenced by the pleiotropic role of programmed cell death in immunity and development? In: Bourtzis K, Miller TA (eds). Insect Symbiosis, vol. 3. CRC Press: Boca Raton. pp 57–75.
Weeks AR, Breeuwer JAJ (2001). Wolbachia-induced parthenogenesis in a genus of phytophagous mites. Proc R Soc Lond B Biol Sci 268: 2245–2251.
Werren JH, Windsor D, Guo L (1995). Distribution of Wolbachia among neotropical arthropods. Proc R Soc Lond B Biol Sci 262: 197–204.
Zchori-Fein E, Faktor O, Zeidan M, Gottlieb Y, Czosnek H, Rosen D (1995). Parthenogenesis-inducing microorganisms in Aphytis (Hymenoptera: Aphelinidae). Ins Mol Biol 4: 173–178.
Acknowledgements
We thank Len Nunney, Roger Butlin, and three anonymous reviewers for helpful comments on the paper. This work was supported by funds from the US National Science Foundation Grant EF 0328363 to RS.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Heredity website
Supplementary information
Rights and permissions
About this article
Cite this article
Russell, J., Stouthamer, R. The genetics and evolution of obligate reproductive parasitism in Trichogramma pretiosum infected with parthenogenesis-inducing Wolbachia. Heredity 106, 58–67 (2011). https://doi.org/10.1038/hdy.2010.48
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2010.48
Keywords
This article is cited by
-
Effects of temperature and superparasitism on quality and characteristics of thelytokous Wolbachia-infected Trichogramma dendrolimi Matsumura (Hymenoptera: Trichogrammatidae) during mass rearing
Scientific Reports (2019)
-
Comparative genomics of the miniature wasp and pest control agent Trichogramma pretiosum
BMC Biology (2018)
-
The effects of outbreeding on a parasitoid wasp fixed for infection with a parthenogenesis-inducing Wolbachia symbiont
Heredity (2017)
-
Nicosulfuron Plus Atrazine Herbicides and Trichogrammatidae (Hymenoptera) in No-Choice Test: Selectivity and Hormesis
Bulletin of Environmental Contamination and Toxicology (2017)
-
Genetics of decayed sexual traits in a parasitoid wasp with endosymbiont-induced asexuality
Heredity (2014)