Abstract
The Chatham Island Taiko (Tchaik, Pterodroma magentae) is one of the world's rarest seabirds. In the past there were millions of breeding pairs of Taiko and it was the most abundant burrowing petrel on Chatham Island. The present population consists of just 120–150 birds, including only 8–15 breeding pairs. Surprisingly high genetic variation was revealed by DNA sequencing of almost every known adult Taiko (N=90). Given the massive population decline, genetic variation may have been even larger in the past. Therefore, we investigated past genetic diversity by sequencing regions of the mitochondrial cytochrome b gene in 44 ancient Taiko bones. We identified a total of 12 haplotypes in Taiko. Eight haplotypes were revealed in the ancient DNA: four were unique to the bones and four corresponded to those found in the modern Taiko population. Surprisingly, despite the critically endangered status of the Taiko, no significant reduction in mitochondrial DNA haplotype diversity was observed between ancient samples (N=44) and modern adult Taiko (N=90). The modern population may have however lost four haplotypes present in the ancient populations.
Similar content being viewed by others
Introduction
A reduction in the genetic diversity of a species can be the result of a severe decline in population size (Spielman et al., 2004). The best method of investigating changes in genetic variation over time is to use ancient DNA (aDNA) derived from samples of the population before it declined. This then provides a direct reference point (Wisely et al., 2002). Using such an approach, studies have compared diversity in pre-bottleneck samples to ‘modern’ samples (for example Chan et al., 2005; Leonard et al., 2005). Some studies have discovered that the change in genetic diversity over a suspected bottleneck was less than expected (for example Paxinos et al., 2002; Miller and Waits, 2003). Such results could only have been obtained by an examination of aDNA.
The critically endangered Chatham Island Taiko (Tchaik, Pterodroma magentae) is one of the world's rarest seabirds (IUCN, 2007). The Taiko is only ever known to have bred on the main island in the Chatham group and nowhere else in the world (Figure 1; Aikman and Miskelly, 2004). The Taiko population consists of just 120–150 birds, including only 8–15 breeding pairs. Surprisingly, genetic variation in the modern adult Taiko population is relatively high in comparison to seabirds and other avian species of varying threat status (Lawrence et al., 2008b). The study by Lawrence et al. (2008b) includes almost every adult Taiko caught and every chick fledged over a 10-year period. There are 10 haplotypes in adult Taiko for the complete cytochrome b gene and 21 for duplicated fragments of the mitochondrial control region domain I. The level of variation of multilocus minisatellite DNA profiles is also high. However, this high level of variation is not present in the generation of chicks born since intense predator control began in 1993 (Lawrence et al., 2008b).
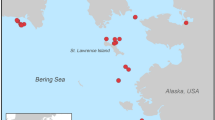
Possible Taiko distribution. Sites where subfossil Taiko bones were found by others (in yellow; Crockett, 1994) and in this study (green). Current Taiko burrow locations (red) and extinct colonies are shown (purple; Sutton, 1979a; DE Crockett, personal communication). Possible extinct Taiko colonies identified by Millener (1999) are indicated by grey circles.
Before the introduction of predators, there were millions of breeding pairs of Taiko and it was the most common of the burrowing petrels on Chatham Island (Aikman and Miskelly, 2004). Given the severe reduction in Taiko numbers (to just 120–150), genetic variation in the species may have been even larger in the past. Past diversity can be investigated using aDNA from widely distributed Taiko bone samples.
Taiko bones have been discovered from many locations around the main Chatham Island, including the northern and eastern regions, primarily from sand dunes (Figure 1; Crockett, 1994). However, many inland areas are unsuitable for bone preservation, due to acidity, peat deposits (and fires therein) and widespread swamps (Millener, 1996). Dune bones could be remains from breeding populations or from beach-wrecked birds (that is birds washed up on the shore; Worthy and Holdaway, 2002), skua (Catharacta lonnbergi) refuse deposits (GA Taylor, personal communication) or birds transported by Moriori (Millener, 1999). Moriori (the indigenous people of Chatham Island/Rekohu) harvested juvenile Taiko from breeding burrows for food (Sutton, 1979a). Birds were preserved for later consumption and may have been transported around the island (Crockett, 1994). Consequently, the presence of juvenile Taiko bones in a location does not necessarily indicate a historical breeding site.
Taiko were an important food source for Moriori living at the Waihora village in the 16th century (Figure 1; Sutton, 1980). Taiko bones make up half of the species assemblages uncovered at some midden sites (human refuse deposits). These sites were likely to be highly specialized seasonal stations that were located near a Taiko breeding colony, within 2 km of the village (Figure 1; Sutton, 1979a). Inhabitants of Waihora were mostly self-sufficient, but there is evidence for some exchange with people from other areas for tools (Sutton, 1982) and perhaps eels, lampreys and inanga (Galaxiid species; Sutton, 1989). Therefore, it is possible that Taiko were used in exchange and transported further north, but evidence is lacking.
The southwest of Chatham Island is generally accepted as the main Taiko breeding area (Crockett, 1994). Crockett identified the locations of Taiko colonies in the southwest of Chatham Island that were very large in the past—of which only one remains today (Figure 1; DE Crockett, personal communication). Nestling bones found with eggshell fragments suggest past Taiko breeding colonies at Long Beach (Figure 1; Millener, 1999). Taiko bones (and in some cases complete skeletons) found at Long Beach have been radiocarbon dated to between approximately 2050 and 3300 CAL BP (Millener, 1999), long before humans reached Chatham Island (about 900 years ago; King, 2000).
Mitochondrial DNA from ancient Taiko bones was sequenced to investigate levels of past genetic diversity in Taiko. Ancient DNA was extracted from Taiko bones collected from around Chatham Island and the archaeological sites around Waihora (Figure 1). Mitochondrial DNA haplotypes were determined by sequencing three regions of the cytochrome b gene totalling 311 bp. These were compared to the haplotypes of the modern Taiko population (Lawrence et al., 2008b). This comparison allows investigation of whether the modern Taiko population has retained or lost its past genetic diversity.
Materials and methods
Ancient DNA extraction
In total, 82 Taiko bones were collected from the surface of sand dunes around main Chatham Island (Figure 1) and 10 were obtained from Canterbury Museum, Christchurch. In addition, 12 Taiko bones were obtained from the Otago Museum, Dunedin. These bones were collected from midden sites around Waihora and are approximately 260 to 450 years old (Figure 1; Sutton, 1976; Sutton, 1979b). Sample details can be found in the Supplementary Information.
DNA extraction and PCR set up was performed in a physically isolated dedicated ancient DNA laboratory. Appropriate measures were taken to prevent and monitor contamination, including extraction and PCR-negative controls. The surface of the subfossil bone material was removed using sterile fine-grained sandpaper. A section was cut from the centre of the bone using a Dremel hand tool. Bone fragments of 15 to 400 mg were ground into a fine powder using a sterile mortar and pestle. The bone powder was decalcified with 0.5 M EDTA (pH 8) and digested with proteinase K/DTT/SDS overnight at 50 °C. The samples were extracted twice with phenol and once with chloroform: isoamyl alcohol (25:1); DNA was recovered with Vivaspin 6 Ultrafiltration Spin Columns (VivaScience, Aubagne, France). Forty-two samples that did not initially amplify with PCR were subsequently purified using a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany).
Ancient DNA amplification and sequencing
PCR primers were designed to amplify three regions of cytochrome b (totalling 311 bp) known to be variable in the modern Taiko population: L14863 (Nunn et al., 1996) and HCytB21–55, 5′-GGGGTGGGTAGGTCAATTAAG-3′ (53 bp); LCytB432 and HCytB571 (152 bp; Lawrence et al., 2008b); and LCytB679, 5′-CCCATTCCATCCCTACTTCA-3′ and HCytB780, 5′-TGGGGAGGTGTAATTAATGGA-3′ (106 bp). Amplification from 2 to 4 μl of DNA extract was carried out in a 20 μl volume using Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA) with the following cycling conditions: hot start 94 °C for 2 min, then 10 cycles of 94 °C for 20 s, 55 °C for 30 s, 72 °C for 30 s; then 50 cycles of 94 °C for 20 s, 52 °C for 30 s, 72 °C for 30 s; then a final extension at 72 °C for 10 min. For amplification from some samples the annealing temperatures were reduced to 52 °C and 50 °C in the first and second set of cycles (respectively).
The PCR products were purified using a QIAquick PCR Purification kit (Qiagen) then sequenced on an Applied Biosystems (Foster City, CA, USA) 3730 DNA analyser by the Allan Wilson Centre Genome Service. Sequences were compared to the reference sequence, that is the most common haplotype found in the modern Taiko population, haplotype A. If any base differentiated from the reference sequence at a site not known to be variable in the modern population, a minimum of two additional PCR amplifications were carried out and sequenced. Miscoding lesions and sequencing artefacts were identified and excluded. Sequences were aligned using Sequencher version 4.2.2 (GeneCodes, Ann Arbor, MI, USA) and deposited in GenBank: accession numbers EU794335–EU794378.
Authentication
DNA from three Taiko bones was independently extracted, amplified and purified for sequencing in another dedicated aDNA facility at the University of Auckland. DNA was extracted using the method described in Matisoo-Smith et al. (1997); the PCR protocol was as described above.
Analysis
A haplotype network was constructed manually. Nucleotide and haplotype diversity was calculated according to Nei (1987) using DnaSP version 4.10.9 (Rozas et al., 2003).
Results and discussion
Mitochondrial DNA haplotype distribution and diversity
DNA was successfully extracted, amplified and sequenced for all three cytochrome b regions from a total of 44 out of 104 bones (42%). These regions identify eight haplotypes in the modern adult Taiko population (Lawrence et al., 2008b; note haplotypes G, H and I are indistinguishable using these three smaller regions). Four of these eight modern adult haplotypes were found in the ancient Taiko bones, in addition to four new haplotypes (Table 1). Each new haplotype is unique, that is found in only one individual and is defined by a single substitution (Table 1; Figure 2). There may have been sites in cytochrome b regions not included in this study that were variable only in the ancient samples and not in the modern samples. This would bias the results towards a conservative conclusion that no change in genetic diversity has occurred.
Novel haplotypes were found in Taiko bones from the central west coast of Chatham Island (one at Paritu and two at Te One; Figure 2). In addition, a unique haplotype was discovered in a bone from Kaingaroa in the northeast (Figure 2). The haplotype network is star-like, with almost all rare haplotypes radiating from common haplotypes (A and G/H/I) and differing from them by a single substitution (Figure 2). This pattern can be indicative of a past population expansion (Slatkin and Hudson, 1991). The haplotype diversity for the 44 aDNA sequences was 0.580 (±0.072 s.d). The equivalent haplotype diversity (that is for the 311 bp) in modern adult Taiko (N=90) was 0.678 (±0.047 s.d). and 0.653 (±0.048 s.d). for the modern Taiko chicks (N=66). Therefore, the range of haplotype diversity overlaps and is not significantly different. Nucleotide diversities were 0.00323 (±3.3 × 10−4 s.d.) for modern adults, 0.00283 (±3.2 × 10−4 s.d.) for modern chicks and 0.00231 (±4.1 × 10−4 s.d.) for aDNA sequences (N=44). The lower nucleotide diversity of the ancient samples suggests there were no post-mortem lesions causing artifactual mutation.
Haplotype diversity over time
Taiko were very numerous in the past, so a higher genetic diversity than that of the very small modern population could be expected. However, the haplotype diversity of the modern adult population (N=90) is not significantly different to that of the ancient bones (N=44). This supports the theory that the modern Taiko population has retained a large proportion of past diversity (Lawrence et al., 2008b). However, some variation may have been lost as four haplotypes present in the ancient populations are not represented in the modern population (which was comprehensively sampled; Lawrence et al., 2008b). The ancient samples used in this study were a mixture of bones hundreds to perhaps thousands of years old, so the Taiko population has retained a reasonably high level of genetic variation for a long period of time. However, the decline in the number of mitochondrial DNA haplotypes in the Taiko chicks over just one generation (that is from eight in the adults to six in the chicks) suggests the Taiko population will lose some of its mitochondrial variation in a very short period (Lawrence et al., 2008b). The level of genetic variation in Taiko maintained over potentially thousands of years could now be lost in a matter of decades.
These results are relevant to Taiko conservation. One of the haplotypes not represented in Taiko chicks (haplotype J; Table 1) is only found in adult males, so cannot be passed on. Another haplotype (D; Table 1) is present in three adult females not known to be breeding. One inhabits a burrow outside of the predator-controlled area and the burrow location of the other two females is unknown. Any chicks these females were to produce would be extremely vulnerable to predation. It is therefore important for Taiko conservation that these burrows be found and protected from introduced predators (Lawrence et al., 2008a).
Accession codes
References
Aikman H, Miskelly C (2004). Birds of the Chatham Islands. Department of Conservation: Wellington.
Chan YL, Lacey EA, Pearson OP, Hadly EA (2005). Ancient DNA reveals Holocene loss of genetic diversity in a South American rodent. Biol Lett 1: 423–426.
Crockett DE (1994). Rediscovery of Chatham Island Taiko Pterodroma magentae. Notornis (supplement) 41: 49–60.
IUCN (World Conservation Union) (2007). IUCN Red List of Threatened Species. IUCN: Gland. Available fromhttp://www.iucnredlist.org(accessed November 2007).
King M (2000). Moriori A People Rediscovered, 2nd edn. Penguin Books (NZ) Ltd: Auckland.
Lawrence HA, Taylor GA, Crockett DE, Millar CD, Lambert DM (2008a). New genetic approach to detecting individuals of rare and endangered species. Cons Biol (in press).
Lawrence HA, Taylor GA, Millar CD, Lambert DM (2008b). High mitochondrial and nuclear genetic diversity in one of the world's most endangered seabirds, the Chatham Island Taiko (Pterodroma magentae). Conserv Genet 9: 1293–1301.
Leonard JA, Vila C, Wayne RK (2005). Legacy lost: genetic variability and population size of extirpated US grey wolves (Canis lupus). Mol Ecol 14: 9–17.
Matisoo-Smith E, Allen JS, Ladefoged TN, Roberts RM, Lambert DM (1997). Ancient DNA from Polynesian rats: extraction, amplification and sequence from single small bones. Electrophoresis 18: 1534–1537.
Millener PR (1996). Extinct birds. In: The Chatham Islands Heritage and Conservation. Canterbury University Press: Christchurch. pp 113–120.
Millener PR (1999). The history of the Chatham Islands' bird fauna of the last 7000 years—a chronicle of change and extinction. Smithsonian Contrib Paleobiol 89: 85–109.
Miller CR, Waits LP (2003). The history of effective population size and genetic diversity in the Yellowstone grizzly (Ursus arctos): implications for conservation. Proc Natl Acad Sci USA 100: 4334–4339.
Nei M (1987). Molecular Evolutionary Genetics. Columbia University Press: New York.
Nunn GB, Cooper J, Jouventin P, Robertson CJR, Robertson GG (1996). Evolutionary relationships among extant albatrosses (Procellariiformes: Diomedeidae) established from complete cytochrome-b gene sequences. Auk 113: 784–801.
Paxinos EE, James HF, Olson SL, Ballou JD, Leonard JA, Fleischer RC (2002). Prehistoric decline of genetic diversity in the Nene. Science 296: 1827.
Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R (2003). DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497.
Slatkin M, Hudson RR (1991). Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 129: 555–562.
Spielman D, Brook BW, Frankham R (2004). Most species are not driven to extinction before genetic factors impact them. Proc Natl Acad Sci USA 101: 15261–15264.
Sutton DG (1976). Radiocarbon dates from the Waihora mound site, south-west coast, Chatham Islands. NZ Archaeol Assoc Newsl 19: 195–196.
Sutton DG (1979a). Island and coastal fowling strategies of the prehistoric Moriori. In: Anderson A (ed). Birds of a Feather Osteological and Archaeological Papers from the South Pacific in Honour of R.J. Scarlett. British Archaeological Reports International Series, vol 62. British Archaeological Reports: Oxford. pp 123–139.
Sutton DG (1979b). Polynesian coastal hunters in the subantarctic zone: a case for the recognition of convergent cultural adaptation. PhD Thesis, University of Otago.
Sutton DG (1980). A culture history of the Chatham Islands. J Polyn Soc 89: 67–93.
Sutton DG (1982). The Chatham Islands. In: Prickett N (ed). The First Thousand Years, Regional Perspectives in New Zealand Archaeology, New Zealand Archaeological Association Monograph 13. Dunmore Press Ltd: Palmerston North. pp 160–178.
Sutton DG (1989). Moriori fishing: intensive exploitation of the inshore zone. In: Sutton DG (ed). Saying So Doesn't Make It So, New Zealand Archaeological Association Monograph. New Zealand Archaeological Association: Dunedin. pp 116–131.
Wisely SM, Buskirk SW, Fleming MA, McDonald DB, Ostrander EA (2002). Genetic diversity and fitness in Black-footed ferrets before and during a bottleneck. J Hered 93: 231–237.
Worthy TH, Holdaway RN (2002). The Lost World of the Moa Prehistoric Life of New Zealand. Canterbury University Press: Christchurch.
Acknowledgements
We thank the Hokotehi Moriori Trust, Te Runanga o Wharekauri Rekohu, the Chatham Island Conservation Board and the Department of Conservation (DOC) for permitting us to collect Taiko bones and Ngati Mutunga for considering our proposal. In addition, landowners Murray and Gill Dix, Mick and Ces Lanauze, Jim and Sally Muirson, Alfred Preece, Pat and Jo Tuanui provided permission to collect bones from their property. We acknowledge Graeme Taylor and Kerry Brown for assisting with bone collection and DOC for providing logistical support. The Canterbury Museum and Moira White, Otago Museum provided subfossil bone samples. We thank Judith Robins for replicating the extraction and amplification of Taiko bones at the University of Auckland. We acknowledge Jennifer Hay and Leon Huynen for technical advice and Vivian Ward for providing Chatham Island maps for figures. This research was funded by the Allan Wilson Centre, Massey University and the University of Auckland.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary information
Rights and permissions
About this article
Cite this article
Lawrence, H., Scofield, R., Crockett, D. et al. Ancient genetic variation in one of the world's rarest seabirds. Heredity 101, 543–547 (2008). https://doi.org/10.1038/hdy.2008.85
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2008.85
Keywords
This article is cited by
-
Ancient DNA studies: new perspectives on old samples
Genetics Selection Evolution (2012)
-
Inferring the ancient population structure of the vulnerable albatross Phoebastria albatrus, combining ancient DNA, stable isotope, and morphometric analyses of archaeological samples
Conservation Genetics (2012)
-
Effects of long-term isolation on genetic variation and within-island population genetic structure in Christmas Island (Indian Ocean) seabirds
Conservation Genetics (2012)