Abstract
Paternity analysis based on microsatellite marker genotyping was used to infer contemporary genetic connectivity by pollen of three population remnants of the wind-pollinated, wind-dispersed tree Fraxinus excelsior, in a deforested Scottish landscape. By deterministically accounting for genotyping error and comparing a range of assignment methods, individual-based paternity assignments were used to derive population-level estimates of gene flow. Pollen immigration into a 300 ha landscape represents between 43 and 68% of effective pollination, mostly depending on assignment method. Individual male reproductive success is unequal, with 31 of 48 trees fertilizing one seed or more, but only three trees fertilizing more than ten seeds. Spatial analysis suggests a fat-tailed pollen dispersal curve with 85% of detected pollination occurring within 100 m, and 15% spreading between 300 and 1900 m from the source. Identification of immigrating pollen sourced from two neighbouring remnants indicates further effective dispersal at 2900 m. Pollen exchange among remnants is driven by population size rather than geographic distance, with larger remnants acting predominantly as pollen donors, and smaller remnants as pollen recipients. Enhanced wind dispersal of pollen in a barren landscape ensures that the seed produced within the catchment includes genetic material from a wide geographic area. However, gene flow estimates based on analysis of non-dispersed seeds were shown to underestimate realized gene immigration into the remnants by a factor of two suggesting that predictive landscape conservation requires integrated estimates of post-recruitment gene flow occurring via both pollen and seed.
Similar content being viewed by others
Introduction
The accuracy of our prediction of the response of forest trees to deforestation and population fragmentation relies on an understanding of how pollen and seed movement is modified as a consequence of changes in the landscape (Sork and Smouse, 2006). Trees, which are characterized by their individual longevity, high intra-population genetic diversity, and often substantial potential for gene flow via pollen and seed, may be particularly well equipped to withstand habitat disturbance (Hamrick, 2004). Although theoretical predictions of reduced genetic diversity and elevated inbreeding following habitat fragmentation (Young et al., 1996) are upheld for a number of wind-pollinated temperate tree species (Sork et al., 2002; Jump and Penuelas, 2006), a recent review of empirical studies conducted in neotropical tree species suggests that fragmentation generally has more complex effects (Lowe et al., 2005).
An emerging picture is of an increase in pollen- and seed-mediated gene flow across deforested landscapes (Aldrich and Hamrick, 1998; Dick, 2001; White et al., 2002; Bittencourt and Sebbenn, 2007). However this enhanced gene flow does not necessarily lead to an increase in genetic diversity or reduction in inbreeding if a limited number of pollen and seed sources contribute to the gene pool (Aldrich and Hamrick, 1998; Sork et al., 2002; O’Connell et al., 2006). Moreover the effect of fragmentation is often not uniform over the landscape. Smaller fragments tend to receive proportionally more pollen immigration than larger fragments because of a paucity of local pollen donors (Sork and Smouse, 2006). It is clear from these considerations that to understand the genetic connectivity of tree species living in fragmented habitats requires an appreciation of the contemporary processes of dispersal and establishment and an analysis of how they are affected by the spatial scale of fragmentation and the heterogeneity of the landscape in which the fragmentation occurs (Sork and Smouse, 2006).
The combined development of highly polymorphic microsatellite markers and statistical analysis of parentage assignment (reviewed in Jones and Ardren, 2003) have made it possible to gather empirical evidence of contemporary gene movement within various landscapes for wild animals (Hazlitt et al., 2006) and plants (Bittencourt and Sebbenn, 2007). For measuring contemporary gene flow these methods have many advantages over previous approaches that relied on inferences from genetic structure and yielded estimates of historical, average values of effective migration rate (Sork et al., 1999; Whitlock and McCauley, 1999). However, a number of problems are beginning to emerge with adopting parentage assignment approaches for measuring gene flow in practice.
The first issue is that the microsatellite methodology widely used for genotyping has significant assay limitations that may call the accuracy of the pedigree inference into question (Dakin and Avise, 2004; Hoffman and Amos, 2005; Slavov et al., 2005). Two main types of genotyping error can be distinguished, allele dropouts (Dakin and Avise, 2004) and erroneous calling of allele size (Amos et al., 2007), both of which can be either of a systematic or stochastic nature. Although some studies suggest that it is best to discard affected loci from parentage analyses where occurrence of non-amplifying (null) alleles is suspected (Dakin and Avise, 2004) Wagner et al. (2006)) argue that when the number of loci is low, discriminatory power may decrease dramatically as a result. They suggest that a better alternative to either removing loci or ignoring the presence of null alleles is to accommodate them within the analyses. Indeed the use of many, even moderately variable, loci rather than fewer hypervariable ones, reduces the impact of error at any particular locus on parentage assignment (Hoffman and Amos, 2005; Slavov et al., 2005). Despite the profusion of recent publications establishing that even a low genotyping error rate had non-trivial consequences for parentage and relatedness studies, quantification and publication of error rates are not yet routinely performed (Hoffman and Amos, 2005).
The second issue is that conclusions drawn from the analysis depend on the method of paternity assignment adopted and assumptions about the size and genetic composition of the population of potential paternal parents that have not been sampled (Oddou-Muratorio et al., 2003; Burczyk and Chybicki, 2004). Although simple exclusion (SE) is a useful starting point for paternity inference, refined statistical approaches are necessary to assess the confidence in paternity assignment (Marshall et al., 1998). For example in natural tree populations it is virtually impossible to sample all trees contributing to the reproductive pollen pool. It is therefore necessary to assess the risk of excluding a candidate pollen parent on the sole grounds that it has not been sampled. Recent methods based on either likelihood or Bayesian approximation allow us to estimate the statistical precision of a paternity assignment for a given sample of a reproductive population (Marshall et al., 1998; Nielsen et al., 2001; Gerber et al., 2003; Araki and Blouin, 2005; Hadfield et al., 2006). Overall, it is preferable to use more than one of these approaches to estimate genetic exchange among populations (Oddou-Muratorio et al., 2003).
In plants a further complication with measuring inter-population gene flow using parentage assignment arises because gene flow is effected by two asynchronous dispersal processes, the first involving pollen and the second involving seeds. The genetic material transferred between populations via pollen and incorporated into seed present on a mother tree only brings about gene flow if that seed is recruited into the local population. Parentage assignment of recruited seedlings is difficult to track with current analytical tools (Sork and Smouse, 2006). Therefore contemporary gene flow among fragmented tree populations has often been estimated by measuring pollen movement in pre-recruitment seeds. These estimates of gene flow are reasonable if seed-mediated gene flow among existing populations is rare, and seed dispersal is primarily important for re-colonization and range expansion. However, in tree species with a high potential for long-distance seed dispersal and subsequent recruitment this assumption may be invalid (Smouse and Sork, 2004) and realized gene flow following seed dispersal may differ significantly from gene flow measured in pre-recruitment seed within a population. The extent of the discrepancy between gene flow measured from parentage analysis before and after seed recruitment has still to be properly documented.
The overall objective of the current study is to describe the genetic connectivity among population fragments of common ash (Fraxinus excelsior) in a chronically deforested landscape in the southern uplands of Scotland. Previous work on these fragments has shown that they maintain high levels of genetic diversity and weak inter-fragment differentiation (Θ=0.080), indicating that historical gene flow has not been limited (Nm=3.48). We also found from an analysis of seed families, using a mixed-mating model approach (Ritland, 2002), that contemporary matings are, on average, predominantly outcrossed (tm=0.971±0.028) and using a neighbourhood model approach (Burczyk et al., 2002) that contemporary effective pollen dispersal distance within the landscape averages 328 m (Bacles et al., 2005). Both seeds collected from forest fragments and newly recruited seedlings were found to harbour high levels of genetic diversity comparable to that of the adult population suggesting an essential contribution of long-distance dispersal to genetic diversity in this wind-pollinated, wind-dispersed species (Bacles et al., 2005, 2006). This paper complements these studies by quantifying the genetic exchange among the individual fragments brought about by pollen flow and relates this to the size and landscape context of the fragments.
To achieve this we estimate pollen-mediated gene flow and male reproductive success of local F. excelsior trees from a paternity analysis of non-dispersed seeds genotyped at hypervariable microsatellite markers. This is the best methodological approach currently available to address this question. Nonetheless, in full awareness of the potential limitations of the methodology, we take a number of steps to ensure that our estimation describes true biological phenomena. First, we quantify genotyping error at marker loci and set out to minimize error because of possible mis-scoring or null alleles using a simple deterministic approach. Second, we use a range of paternity assignment methods to obtain a confidence interval rather than a point estimate of pollen-mediated gene flow. Finally, we compare such estimates derived from non-dispersed seeds with those derived from seedlings establishing in the same F. excelsior remnants estimated by means of parent-pair analysis (Bacles et al., 2006), to assess how they relate to absolute levels of genetic exchange among remnant populations.
Material and methods
Study species
F. excelsior, common ash, is a post-pioneer tree species widespread in temperate Europe and native throughout the British Isles. The phylogeography of the species is now well described (Morand et al., 2002; Heuertz et al., 2004; Ferrazzini et al., 2007). F. excelsior displays a complex, polygamous sexual system (FRAXIGEN, 2005) in which individuals may be classified phenotypically across a continuum from purely male to purely female with a whole range of hermaphroditic intermediates. Hermaphrodite individuals are self-fertile and levels of seed sets are similar in hermaphrodite and female trees, but in natural populations, F. excelsior is preferentially outcrossed and male fertility of hermaphrodite trees appears to be much lower than that of male trees (FRAXIGEN, 2005). Fruits are dry and winged, adapted to wind dispersal. Regular fruit bearing begins around 20 years of age but fruiting phenology will vary depending on latitude, altitude, temperature and between years with great variation from no seeding to masting (FRAXIGEN, 2005).
Sampling and data collection
The study site is a highly deforested catchment of 900 ha (Moffat Dale) located 80 km south of Edinburgh, Scotland (N 55° 23′ 51′′, W 3° 19′ 50′′), which forms part of a glacially derived landscape in which steep-sided valleys have been carved by ice. Many native tree species including F. excelsior are confined to steep and narrow streamsides situated at the bottom of steep valleys inaccessible to grazing herbivores. Populations of F. excelsior tend to be very small, comprising 10–30 mature individuals, with no natural regeneration in grazed areas. Remnant stands are typically separated by hundreds of metres although some can be isolated by more than one kilometre. In this catchment, F. excelsior is present in only five forest remnants, two of them within the Carrifran Burn and three others in its immediate surroundings (Figure 1; see also Bacles et al., 2005).
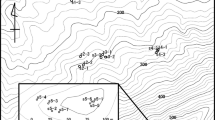
Distribution of Fraxinus excelsior mature trees in Moffat Dale remnants. Each dot represents a tree. (A) Mature trees grow in small forest remnants, confined to steep slopes (elevations are given in meters) along streams (highlighted in dark lines). An exhaustive sampling and mapping was performed in remnants CMa (N=4), CDa (N=30) and SCa (N=12) whereas in two larger remnants SBa and Wa, which include approximately 50 mature trees each, 20 individuals were sampled throughout each of them as potential sources of immigrant pollen flowing into remnants CMa, CDa and SCa. Two lone trees of the Carrifran Burn (labelled A and B) were also sampled. (B–F) Close-up of spatial distribution of individuals sampled in remnant CDa, CMa, SCa, SBa and Wa respectively. In CDa, CMa, and SCa, all individuals producing fruits in 2000 are represented by a star: 30 seeds were collected throughout the tree canopy from 11, 2 and 6 trees in CDa, CMa and SCa respectively. The background map is a section of Ordnance Survey product Land-line.Plus-nt11 Crown copyright Ordnance Survey. An EDINA digimap/JISC supplied service. Figure 1 was originally published in Bacles et al. (2005) reprinted with kind permission from Evolution (Blackwell Publishing).
Two remnants in the bare open landscape of the Carrifran Burn, CDa and CMa, and one remnant confined to a higher altitude dense conifer plantation upstream of Carrifran in Swine Cleuchs, SCa, chosen for their heterogeneity in size, density and landscape features, were exhaustively sampled for adult trees and family arrays. Two neighbouring remnants, in Spoon Burn (SBa), the adjacent valley to Moffat Dale nearest to Carrifran, downstream and in Whitewells (Wa) located at the bottom of the Moffat Dale where the Carrifran streams drain into Moffat Water (Figure 1), were partially sampled for adult trees to gather an indication of potential pollen immigration because they are the only two other known local pollen sources within 10 km.
In 2000 and 2001, leaf material was collected from all mature trees in CDa, CMa and SCa (comprising 30, 4 and 12 individuals respectively; Figure 1). Leaf material was also collected from two trees (A and B) isolated from the nearest remnants by a distance of 250 m and from a sample of 20 mature trees in SBa and Wa (Figure 1). Such sampling represents no less than 40% of the composition of these two remnants. Thirty fruits, or all fruits if the seed crop was less, were collected from all 19 trees producing fruits in 2000, a non-masting year, in each of remnants CDa, CMa and SCa. In total, we sampled 88 trees and 483 seeds from 19 families.
The complete sample of 88 trees and 483 seeds was genotyped for five microsatellite markers previously developed for F. excelsior, namely, M2-30B, 1.19 and 3.1 (Brachet et al., 1999), and FEMSATL2 and FEMSATL5 (Lefort et al., 1999). DNA isolation, amplification by polymerase chain reaction (PCR) and electrophoretic separation of PCR products were carried out as described elsewhere (Bacles et al., 2005).
Evaluating microsatellite scoring and accounting for mistyping
Out of 483 seeds genotyped, 61 presented a mismatch with their mother at one or more loci. Furthermore, genotypes observed at loci 3.1 and FEMSATL5 suggest departure from Mendelian segregation (Bacles et al., 2005) and occurrence of null alleles that has also been discussed in other F. excelsior studies (Heuertz et al., 2001; Morand et al., 2002).
In the rare instances where correction for genotyping error is applied in empirical studies, it is generally introduced as a global stochastic error rate (Marshall et al., 1998; Gerber et al., 2000). A major drawback of such practice is that the benefits of accounting for error are often outweighed by costs in precision of paternity assignment that becomes uninformative (Oddou-Muratorio et al., 2003; Morissey and Wilson, 2005). Therefore, here we chose to account for allele dropouts and size miscalls deterministically by performing two successive transformations to the raw multilocus genotypes (referred to hereafter as RAW).
Erroneous allele sizing is most likely to occur between alleles of similar size and when alleles are rare. Therefore, we applied an initial transformation to account for size miscalls in the form of a binning procedure. At each locus, rare alleles were binned with common alleles of the nearest size. Alleles were deemed rare when they occurred at a frequency of less than 0.01 for the entire data set. The procedure was applied strictly to loci M2.30B and FEMSATL2. For loci 1.19, 3.1 and FEMSATL5, more common alleles were also binned to reflect difficulties in gel scoring for 1.19 and difficulties in respect of Mendelian segregation for 3.1 and FEMSATL5 (Bacles et al., 2005). The procedure reduced the number of alleles observed from 54 to 29, 35 to 20, 10 to 7, 46 to 12 and 17 to 8 at loci FEMSATL2, M2-30B, 1.19, FEMSATL5 and 3.1 respectively in the binned data set (referred to hereafter as BIN).
A second transformation was then performed to account for the possible occurrence of allele dropouts. A one-allele dropout model was applied to each locus by introducing a new (that is, non-observed) allele, by re-scoring every individual with a non-amplifying genotype as homozygote null, and every observed homozygote as heterozygote null in the transformed data set (hereafter referred to as BINNULL).
For each data set, genotyping error rates were quantified by means of direct comparison of offspring–mother genotype at each locus and averaged over loci in CERVUS 2.0 (Marshall et al., 1998). Loci were retained for subsequent analyses if the estimated genotyping error was less than 5%. To assess the discriminatory power of each data set, a paternity exclusion probability (PEP) was computed for each locus and accumulated over loci in FAMOZ (version released on 17 April 2007; Gerber et al., 2003).
Estimating contemporary pollen-mediated gene flow at the landscape scale
Paternity analyses were undertaken to identify the pollen parent of the 422 seeds that shared a compatible multilocus genotype with their putative mother only and excluding the 61 seeds presenting at least one mismatching allele with their mother. Pollen parents were considered either among the 48 trees sampled in CDa, CMa and SCa, including the possibility for mother trees to self, or as pollen-mediated gene flow from outside the landscape covered by the three completely censed remnants (Figure 1).
For each of the RAW, BIN and BINNULL data sets, paternity was assigned using both a SE and a maximum-likelihood (ML) approach in FAMOZ (Gerber et al., 2003). In each case, outcomes of paternity assignment may be, for each individual seed, either that one unique individual among the 48 trees of Carrifran and Swine Cleuchs is assigned as its pollen parent, or that its paternity is unresolved with more than one possible pollen parent among the 48 trees, or finally that all 48 trees are excluded as potential pollen parents and its paternity is assigned to immigrant pollen. A range of values for apparent pollen-mediated gene flow into the landscape is subsequently obtained as the percentage of seeds in the sample for which paternity was assigned to immigrant pollen.
In FAMOZ, confidence in paternity assignment is estimated using a simulation procedure for hypothesis testing (Gerber et al., 2000). The paternity of the 422 seeds sampled from 19 mother trees in three F. excelsior remnants of Moffat Dale was assigned to the most-likely fathers detected by means of ‘log of the odds’ ratios (LOD scores) based on pollen pool gene frequencies estimated from progeny arrays in MLTR (Ritland, 2002). We chose to approximate the (non-observed) allele frequencies of the reproductive population by using the observed pollen pool frequencies instead of the frequencies observed for the small sample of 88 mature trees because the latter, which is sampled a priori based on spatial location, may be a poor estimate of the actual reproductive population if gene flow is extensive. No significant genotypic association was detected among any pair of loci (Bacles et al., 2005). LOD scores over all loci were therefore obtained by adding LOD scores calculated for each locus.
Confidence in paternity assignment was then determined in FAMOZ by comparing the distribution of the LOD scores of the most-likely fathers of 50 000 randomly generated seeds with their father randomly chosen among the 48 trees to the distribution of LOD scores of the most-likely fathers of 50 000 seeds whose paternal genotype was randomly generated according to pollen pool allele frequencies. The test threshold for rejecting a candidate as a true father (TF) was chosen at the intersection of the two distributions of LOD scores to minimize both type I error, wrongly considering as resulting from pollen immigration a seed sired by a sampled father, and type II error, wrongly assigning true pollen immigration to a sampled father (Gerber et al., 2000). For paternity assignment by SE, all candidate males with a positive LOD score (that is, test threshold TF=0) were not excluded from being the true father.
Global results of paternity assignment obtained for each of RAW, BIN and BINNULL data sets with both SE and ML methods are discussed in respect of estimated error rates, confidence levels in assignments and estimated pollen-mediated gene flow at the landscape scale. The data set/method combination found to minimize genotyping error rates while maximizing confidence in paternity assignments was retained for subsequent detailed description of individual male reproductive success. In particular, results were summarized to identify the number of sires among the sample trees and the number of seeds they sired among the sampled seeds. The pollen dispersal curve was estimated by plotting the distance between mother trees and pollen donors for each most-likely assignment. When more than one likely father was identified (unresolved assignment), a fraction of the seed was assigned to all likely fathers evenly and proportionally to the number of likely fathers found.
Estimating the fractional pollen contribution of forest remnants and identifying local sources of pollen immigration
To estimate landscape connectivity and pollen-mediated genetic exchange among forest remnants, it may be most relevant to assess the relative pollen contribution of forest remnants to the seed crop rather than to define individual paternity per se. It has been argued that such (meta)population scale phenomena may be better addressed with fractional-likelihood (FL) assignment methodology that will assign a fraction of the paternity of a given seed to all male candidates with a positive LOD score in proportion to their likelihood probability (Nielsen et al., 2001). To estimate potential pollen immigration into CDa, CMa and SCa from other known F. excelsior remnants of Moffat Dale, SBa and Wa (Figure 1), we estimated the posterior expectation of the number of sampled offspring in each of five remnants by means of FL assignment in PATRI (Signorovitch and Nielsen, 2002).
The approach in PATRI also allows us to make prior assumptions about the proportion of the pollen parents that have not been sampled (Nielsen et al., 2001). Although the actual effective male population size is unknown, we do have some expectations of the number of trees occurring in the landscape and likely to contribute to the pollen pool. Therefore, we tested the sensitivity of the FL assignment to assumptions made on the population size (N) by successively repeating analyses for an N of 88, the number of trees sampled; an N of 150, the approximate number of F. excelsior trees occurring in the catchment and N modelled as a uniform function varying between 100 and 500. We compared results with those of an ML assignment performed in FAMOZ when considering all 88 trees sampled in Moffat Dale.
Comparing potential to realized pollen-mediated gene flow among forest remnants
To assess whether estimates of pollen-mediated gene flow from seeds that have not yet dispersed reflect estimates of pollen-mediated gene flow seen after seed dispersal and establishment, for each of remnant CDa, CMa and SCa, we used most-likely fathers to attribute the origin of the pollen grain to either local pollen, foreign pollen of known source (in other identified remnants) or of unknown source. We compared these figures with previously published results derived from an ML parent-pair analysis performed on seedlings establishing in the same three remnants (Bacles et al., 2006).
In addition, we estimated total gene flow into fragments using genotypic data generated both from progeny arrays (Tp) and from established seedlings (T′s). Note that pollen grains only carry one gene copy whereas diploid seeds carry two. Let A and A′ represents the number of local seeds fertilized by immigrant pollen in progeny arrays and establishing seedlings respectively. Let B′ be the number of immigrating seeds, and C and C′ the total number of seeds sampled in progeny arrays and establishing seedlings respectively, then:

and

In Equation 1, seeds are sampled on known mother trees and are all local. Tp therefore assumes that pollen is the main vector of gene flow among populations and that seed dispersal is mostly local. Results are discussed in terms of comparison of potential (that is, ante dispersal) and realized (that is, post dispersal and establishment) gene flow in the three heterogeneous forest remnants.
Results
Genotyping error and choice of data set
Genotyping error rates estimated by means of offspring–mother genotype comparison are reported for each locus and each genotype transformation in Table 1. They were found to be highest for loci 3.1 (error (RAW)=0.2847) and FEMSATL5 (error (RAW)=0.2237). Drastically reducing the number of alleles, from 17 to 8 and from 46 to 12 at locus 3.1 and FEMSATL5 respectively, decreases the error rate only slightly (error (3.1, BIN)=0.2679; error (FEMSATL5, BIN)=0.2133) but additional inclusion of a null allele decreases the error significantly (error (3.1, BINNULL)=0.1499; error (FEMSATL5, BINNULL)=0.0762), suggesting that null alleles may be responsible for the non-Mendelian segregation observed in progeny arrays at these loci. In contrast, genotyping error rates estimated at loci 1.19, M2.30B and FEMSATL2 were under 5% (Table 1), and lowest at loci M2.30B and FEMSATL2 after binning of alleles (error (M2.30B, BIN)=0.0347; error (FEMSATL2, BIN)=0.0443) and with inclusion of a null allele at locus 1.19 (error (1.19, BINNULL)=0.0289). Overall, inclusion of loci 3.1 and FEMSATL5 in estimates increases mean error across loci dramatically, up to 3 times (Table 1). When these loci were excluded, estimates of mean error across loci were consistently under 5%.
PEPS estimated per locus for RAW vary between 0.594 at locus 1.19 and 0.864 at locus FEMSATL2 (Table 1) reflecting variation in level and evenness of polymorphism among loci (Bacles et al., 2005). As expected, reducing the number of alleles at each locus systematically lowers discriminatory power among genotypes at each locus, albeit moderately, with PEP estimated per locus for BIN varying from 0.504 at locus 1.19 to 0.857 at locus FEMSATL2. Conversely, additional inclusion of a null allele results in contrasting effects on single locus estimates of PEP (Table 1). However, cumulative estimates of PEP, including all five loci, are consistently very high across data sets, reaching values upward of 99.9%. Excluding loci 3.1 and FEMSATL5 decreased cumulative PEP only slightly (Table 1).
On the basis both that including loci 3.1 and FEMSATL5 increases genotyping error to rates well above 5% and that excluding them hardly affects multilocus genotype discrimination of individuals, they were not retained for subsequent analyses. Meanwhile, considering loci 1.19, M2.30B and FEMSATL2 only, mean genotyping error rate is lowest when multilocus genotypes are transformed with binning at loci M2.30B and FEMSATL2 and with binning and inclusion of a null allele at locus 1.19 (data set hereafter referred to as BIN3NULL1; error (mean, BIN3NULL1)=0.0360). No identical multilocus genotype was found among the 88 trees sampled in Moffat Dale and cumulative PEP is estimated at 0.991, which is nearly as high as for RAW data (Table 1).
Contemporary pollen-mediated gene flow at the landscape scale
Results of SE and ML paternity analysis for RAW, BIN, BIN3NULL1 data sets and for assignment of pollen parents to the 422 seeds that do not show any mismatch with their mother on raw data are given in Table 2. In total 43–68% of the 422 seeds analysed were found to have been fertilized with pollen dispersed from trees located outside the landscape covered by the three F. excelsior remnants of Carrifran and Swine Cleuchs. Highest estimates were obtained using an ML method (Table 2). Differences between SE and ML estimates of apparent pollen flow are due to a number of seeds that were not assigned a father among the 48 sampled trees with the ML method because their LOD was positive but below the given threshold for assignment. However, they were assigned one father (or several potential fathers) with the SE method, most frequently for the bin data set (Nassigned (BIN, SE)=239; Table 2). False rejection of true sampled fathers was lowest (type I error <0.05 for TF=2.90) for fewer seeds (Nassigned=141) with the BIN3NULL1 transformation (Table 2).
Individual male reproductive success and distance of pollen dispersal
On the basis that the BIN3NULL1 data set is characterized by the lowest estimates of genotyping error and type I error in ML assignment, the most-likely pollen parents identified for 141 seeds under these conditions were retained for subsequent description of male reproductive success and spatial patterns of pollen dispersal at the landscape scale.
In total, 31 of the 48 trees sampled in CDa, CMa and SCa were found to sire one or more of the sampled seeds (Supplementary Table S1). Of these, 14 trees fertilized three seeds or less, whereas three trees fertilized more than ten seeds each. In the latter case, all fertilized seeds were sampled from neighbouring trees within the same remnant as the sire.
This pattern in individual male reproductive success is reflected in the L shape of the pollen dispersal curve constructed by plotting the distribution of spatial distances between the 141 assigned seeds (that is, based on spatial location of mother trees) and their most-likely pollen parent (Figure 2a). Over 80% of effective pollen dispersal is confined to less than 100 m from the source, corresponding to local pollen movement within remnant. The observed proportion of these local pollinations is significantly higher than expected under random dispersal (48%, Wilcoxon two-sided signed-rank test, P-value<0.05; Figure 2a). However, a number of rarer events, each representing less than 10% of total effective pollen dispersal (which is up to 25% less than expected from random dispersal), were identified between 200 and 600 m, and between 1600 and 1800 m corresponding to inter-remnant long-distance dispersal.
Comparison of the frequency distribution of possible (white) and detected (black) effective pollen dispersal events for Fraxinus excelsior in relation to the location of pollen donors within the fragmented landscape of Moffat Dale. (a) Detection within three censed remnants. Possible dispersal distances were estimated from Euclidian distances between 141 assignable seeds, spatially located on 19 mother trees, and all 48 F. excelsior candidate pollen parents sampled in remnants CDa, CMa and SCa of Moffat Dale (Figure 1). Detected pollen dispersal distances were estimated from Euclidian distances between the 141 assignable seeds and their likely father when identified by means of maximum-likelihood paternity analysis in FAMOZ (Gerber et al., 2003; Table 2). (b) Detection including neighbouring sources of immigrant pollen. Possible dispersal distances were estimated from Euclidian distances between 163 assignable seeds, spatially located on 19 mother trees, and all 88 F. excelsior candidate pollen parents sampled in all five remnants of Moffat Dale (Figure 1). Detected pollen dispersal distances were estimated from Euclidian distances between the 163 assignable seeds and their likely father when identified by means of maximum-likelihood paternity analysis in FAMOZ (Gerber et al., 2003; Table 3). In both situations, when more than one likely father was identified (unresolved assignment), a fraction of the seed was assigned to all likely fathers evenly and proportionally to the number of likely fathers (Supplementary Tables S1 and S2). Distance distributions of detected pollen dispersal were found to differ significantly from random dispersal (Wilcoxon two-sided signed-rank test, P-value <0.05 for both n=141 and n=163).
Further analysis that included trees in partially sampled remnants SBa and Wa allowed us to refine the shape of the pollen dispersal distribution (Figure 2b). ML paternity assignment of 163 seeds (TF=2.92, type I error <0.05; type II error<0.28) attributed their paternity either to trees sampled in CDa, CMa and SCa or to trees sampled in SBa and Wa (Supplementary Table S2). The inclusion of these nearby pollen sources allowed the detection of a small number of effective pollination events at distances between 1100 and 1600 m, 1800 and 2000 m and as great as 2900 m (Figure 2b).
Pollen-mediated genetic connectivity of forest remnants
Pollen contribution of the five F. excelsior population remnants sampled for adult trees was estimated from FL paternity analysis of a subset of 404 seeds and 83 male candidates with no missing value in their multilocus genotype at 1.19, M2.30B and FEMSATL2 because PATRI computes complete genotypes only (Signorovitch and Nielsen, 2002). Estimates of the relative contribution of the five F. excelsior remnants of Moffat Dale to effective pollination in remnants CDa, CMa and SCa are similar when an FL or ML approach is applied to all 88 genotyped trees (Table 3). However, the absolute fractional contribution of remnants to paternity of sampled seeds is highly sensitive to the sampled fraction of reproductive adults (Nielsen et al., 2001) as illustrated by decreasing posterior expectation of the number of sampled offspring fathered in each remnant with increasing prior N (Table 3). Overall however, the relative pollen contribution of the five forest remnants remains unchanged. CDa contributes most to paternity of the sampled seeds. SCa, SBa and Wa also contribute in decreasing proportion (Table 3), whereas remnant CMa is a poor contributor (Table 3). The analysis clearly demonstrates that neighbouring remnants may act as sources of immigrant pollen.
Estimates of pollen-mediated genetic exchange among remnants CDa, CMa and SCa derived from ML paternity analysis confirm that the largest remnant CDa acts as a pollen donor, siring 26% of the seeds sampled in remnant CMa, located 600 m away, and 7% of the seeds sampled in the most spatially isolated remnant, SCa, located 1700 m away (Figure 3). Conversely, CMa, the smallest remnant (Ntrees=4), only sired 3% of its local seeds, and 1% of the seeds within CDa (Figure 3).
Schematic map of pollen-mediated genetic exchange among three Fraxinus excelsior forest remnants varying in their population size, density and degree of spatial isolation to other forest remnants in the mosaic landscape of Moffat Dale. Estimates of gene movement within remnants (continuous white arrows), of gene flow among remnants (continuous black arrows) and of gene immigration from external sources (dashed white arrows) are based on results of maximum-likelihood (ML) paternity analysis performed in FAMOZ (Gerber et al., 2003); 422 seeds sampled from all 19 seeding trees in remnants CDa (Ntrees=30), CMa (Ntrees=4) and SCa (Ntrees=12), considering all 48 trees occurring within the landscape, including two isolated trees (A, B; Figure 1) as potential pollen donors (Table 2). Relevant potential geographic barriers to gene flow among remnants are highlighted: remnants CDa and CMa are located in close proximity (600 m) at the bottom of the bare and open valley while remnant SCa is most isolated over a ridge (dashed white rectangle) located about 1700 m away and surrounded by a dense closed conifer plantation (continuous black square).
Potential and realized pollen-mediated gene flow
How such pollen-mediated genetic exchange will affect genetic structure depends on dispersal and establishment of the seeds. Estimates of potential pollen-mediated gene flow into remnants CDa, CMa and SCa from ML paternity analysis of 422 seeds collected on mother trees before their dispersal described above (65–94%; Table 4) are comparable to those of potential pollen-mediated gene flow from an ML parent-pair analysis of 60 seedlings that were establishing in the same three remnants the following year (70–100%; Table 4). However, such comparison also shows that pollen-mediated gene flow realized after seed dispersal and seedling establishment is much lower, ranging from 12.5% in remnant CMa to 17.5% in remnants CDa and SCa. Total gene flow estimates from progeny arrays are much lower (Tp ranging between 32.5 and 47%) than from establishing seedlings (T′s ranging between 67.5 and 87.5%; Table 4).
Discussion
Despite a number of significant concerns over genotyping error and uncertainties associated with statistical modelling, the application of paternity assignment analysis in the fragmented populations of F. excelsior has significantly enhanced our understanding of their genetic behaviour. It is clear that the population fragments within a single valley receive about half their pollen from outside the valley. Remnants within the valley are genetically connected via pollen flow, but the patterns of pollen flow among fragments are not symmetrical; pollen is preferentially transferred from large to small fragments. The analysis has also demonstrated that the effective pollen dispersal curve is fat tailed. Although the majority of detected pollen movement occurs over short distances (within 100 m), there is still substantial pollen flow occurring over distances greater than 1 km. Although these general conclusions are important for guiding the management of fragmented tree populations, this study has also highlighted the practical difficulties associated with obtaining quantitative assessments of gene flow from large-scale studies that rely on parentage analysis.
A predictive understanding of the genetic connectivity of fragmented populations requires reliable estimation of contemporary gene dispersal across heterogeneous landscapes (Sork and Smouse, 2006). Although development of both molecular techniques and statistical tools has greatly improved prospects for accuracy, the application of parentage analyses to natural populations remains an evolving area of research leading to regular re-analysis of empirical data within new statistical frameworks (Slate et al., 2000; Hadfield et al., 2006). At the centre of the debate lies the question of sensitivity of parentage analyses to partial sampling of the reproductive population and to genotyping error at marker loci (Nielsen et al., 2001; Oddou-Muratorio et al., 2003; Slavov et al., 2005). To obtain reliable population-level inference of gene flow from a collection of individual-level paternity assignments, we chose to address these concerns by applying a range of paternity analysis methods to F. excelsior population remnants of the chronically deforested catchment of Moffat Dale (Tables 2 and 3).
Critically, application of parentage analyses to estimating gene movement in natural populations relies on a conundrum: accuracy in estimation of the proportion of the reproductive population that has not been sampled (that is, immigrant gene flow) strongly increases as the proportion of the—yet unknown—reproductive population that has not been sampled decreases (Oddou-Muratorio et al., 2003). Approaching true reproductive population size seems particularly important to analyses performed when using a FL approach in PATRI because estimating the absolute contribution of F. excelsior trees to paternity of sampled seeds is sensitive to input prior information on the effective male population size (Table 3). The advantage of the hypothesis testing-based simulation approach to determine assignment confidence in FAMOZ is that it does not require any assumption on the size of the true reproductive population. Comparison of SE and ML methods for a range of transformed multilocus genotypes accounting for genotyping error sensu lato at microsatellite markers suggests an immigration of at least 43% (SE, BIN) and up to 68% (ML, RAW) of the pollen-fertilizing seeds from 19 trees of three forest remnants of the Moffat Dale catchment.
The range in gene flow rates seems mostly affected by the choice of paternity assignment method rather than by data set transformation. Indeed, although transformation of raw data allowed us both to reduce mean genotyping error (for BIN3NULL1 at 3.60%), and to minimize false rejection of TFs that were sampled (for BIN3NULL1 type I<5%) to acceptable levels, estimates of gene flow among RAW, BIN and BIN3NULL1 vary by up to 15% for a given paternity assignment method. Variation in gene flow estimates between the SE and ML methods can be attributed to the fact that under ML between 10 and 16% of seeds are not assigned a father among the sampled trees. This is because the LOD score of candidates is positive but below the given threshold for assignment (TF=2.90; Table 2). Although paternity of these seeds cannot be assigned at the chosen confidence threshold, it is arguable that their paternity should necessarily be attributed to immigrant pollen. Indeed, it has been demonstrated that assignment error may be much higher than random on unobserved (that is, immigrant) events (Slate et al., 2000), which suggests that estimates of pollen-mediated gene flow from ML method (here inclusive of seeds that were not assigned a father because genotypically compatible candidates had a low LOD score) should be seen as upper limits. Conversely, ML analysis suggests that even for a strict LOD score threshold, type II error of wrongly assigning immigrant pollen to an unrelated sampled tree is high (up to 27% for BIN and TF=2.50; Table 2) indicating substantial cryptic gene flow. Therefore, apparent pollen flow estimated with relaxed assignment (equivalent to TF>0) by SE method, and those obtained with BIN data because allele binning results in lower discrimination of multilocus genotypes, are most conservative, with increased risk of cryptic gene flow and therefore represent lower limits of effective pollen immigration into forest remnants of Moffat Dale.
We justify our deterministic transformation of genotypic data not as substitution of raw data sets for transformed ones that may be more biased but rather as a simple way of minimizing genotyping error and its possible influence on paternity assignment. The transformed data set still includes a mean genotyping error rate of about 3.6% per locus, which may still have an impact on the conclusions drawn from this study. Nonetheless, we deliberately chose not to include this global rate in paternity analyses because there is evidence that including global genotyping error rates inflates errors in paternity assignments (Oddou-Muratorio et al., 2003; Slavov et al., 2005). Given the limitations of the data set, which are here clearly quantified, a range of estimates from several paternity analyses provides an ecologically meaningful interpretation of pollen-mediated gene flow at the landscape scale.
Extensive contemporary pollen-mediated gene flow averaging 60% has already been reported in plots located within continuous stands of F. excelsior (Hebel et al., 2007) and of other wind-pollinated temperate tree species, for instance for Quercus (Dow and Ashley, 1998), covering only small areas. Contemporary pollen-mediated gene flow estimates of 43–68% for F. excelsior in the fragmented landscape of Moffat Dale are comparatively higher because all F. excelsior trees were sampled in an area of 300 ha suggesting that F. excelsior maintains extensive pollen exchange across a landscape heavily deforested not only locally but also at the wider regional scale of the southern uplands of Scotland (>50 km). Although trees standing solitarily in grazed pastures, spatially isolated from congeners following deforestation, were once described as living-dead (Janzen, 1986), there is now plethora of evidence of reproductive activity of isolated pasture trees, mainly in tropical species (Aldrich and Hamrick, 1998; Dick, 2001; White et al., 2002) corroborating our findings of enhanced pollen-mediated gene flow following anthropogenic disturbance. Nonetheless, of the two F. excelsior trees of Moffat Dale that are isolated from others by a distance of at least 250 m (Figure 1), neither produced a seed crop nor did they contribute to effective pollination of seeding trees for the sampled reproductive season. On the basis of this observation, we cannot reject the hypothesis that such isolated pasture trees are living-dead. Similarly, only three of the 48 trees sampled locally have a high male reproductive success and 26 of them contribute to effective pollination of fewer than two seeds to none (Supplementary Table S1). However, whereas the evidence suggests that most sampled trees have a low individual male reproductive success locally, we cannot reject either the hypothesis that pollen from such trees effectively emigrated to other forest remnants outside the sample area, as the presence of a large component of immigrant pollination of either unknown origin, or originated from identified neighbouring sources would suggest (Figure 3).
Several ecological factors may have contributed to confer such an advantage to long-distance pollination. First, comparison of temporal variation of effective pollen movement between mast seeding and non-masting years showed that in a non-masting year, as is the case in the present study, pollen-mediated gene flow was favoured in a F. excelsior stand in southern England (FRAXIGEN, 2005). Furthermore, in such a situation, the small seed crops that were produced by a number of trees (in particular, trees SCa34 and SCa38 displayed only one seed branch with fewer than 10 seeds; Supplementary Table S1) may create a sampling effect. Second, temporal variation in individual flowering phenology may greatly affect mate availability. Indeed, Gerard et al. (2006) not only found that co-flowering individuals were patchily distributed in space in a F. angustifolia and F. excelsior hybrid zone, they also detected an asymmetry in male reproductive success with early flowering trees participating more as pollen donors than late flowering ones. A scenario where immigrant pollen would be preferentially available during the period of stigma receptivity of seeding trees of Moffat Dale would also favour long-distance pollination.
However, high levels of gene immigration are not necessarily sufficient to prevent assortative mating and selfing (Gerard et al., 2006). For gene flow to become an efficient force counteracting the deleterious genetic effects of habitat fragmentation, it must not only be sustained at high levels across seasons, but must also be qualitatively diverse. Here we find that efficient pollen immigration allows for new and diverse genetic material to establish in the seed generation (Bacles et al., 2005). Such genetically diverse pollen pool composition may be explained by the type of decay of pollen dispersal. Indeed, Klein et al. (2006) demonstrated by means of simulation that fat-tailed dispersal kernels lead asymptotically to a diverse propagule pool containing a balance of mixing of the propagules of two sources and therefore that the diversity of the pollen pool of a mother plant should increase with increased spatial isolation. Pollen dispersal patterns observed for F. excelsior in Moffat Dale seem to corroborate such theoretical findings. The majority of detected pollen dispersal was found between near-neighbours at distances under 100 m (Figure 2). However, not only were a number of rare events detected among forest remnants at distances up to 2900 m, in proportions departing from random dispersal (Wilcoxon two-sided signed-ranked test, P<0.05), but undetected events were also in the majority and may have originated at much greater distances, suggesting an L-shaped pollen dispersal kernel with a tail spreading over several kilometres and underlining the difficulty of detecting long-distance dispersal (Nathan, 2006).
Such pollen dispersal effective over long distances can be linked to landscape features resulting from habitat disturbance. Indeed, in the southern uplands of Scotland, deforestation and land use for pasture have greatly opened the barren landscape that is regularly battered by strong winds. It is therefore likely that wind-mediated pollen movement for F. excelsior has been facilitated by the modification of the landscape in Moffat Dale. In particular, genetic connectivity of forest remnants seems favoured by landscape openness and remnant size rather than by geographic proximity. Indeed, although no seeding trees were sampled in two of five remnants (SBa and Wa), FL paternity assignment shows that their contribution to effective pollination of seeding trees of the other three extant remnants (CDa, CMa and SCa) is higher than that of remnant CMa which is a much smaller remnant of only four trees located in the bed of the river running through an exposed and barren pasture in the Carrifran valley. Within-remnant pollination for CMa is indeed much lower than for other remnants (Figure 3), highlighting the fact that remnants are smaller, but are spatially well connected to neighbouring forest remnants, and tend to receive proportionally higher gene flow simply because there are fewer potential local pollen donors (Sork and Smouse, 2006).
An important practical consequence of the high rate of pollination by immigrant pollen is that the locally produced seed in Carrifran will contain genes sampled from a wide geographic area around the valley. How the high rate of success of immigrant pollen in the production of local seeds will ultimately affect the genetic structure of F. excelsior remnants depends on how much of the pollen pool genetic diversity is effectively carried into successive generations by established seedlings that reach maturity. Natural regeneration in Moffat Dale has been severely limited by continuous grazing pressure. Colonization of mountain grasslands by F. excelsior seedlings has been found to be connected to grazing activities, with seedlings found preferentially in high layers of vegetation, in shaded and ungrazed areas (Julien et al., 2006). Pasture habitats in Moffat Dale may therefore be unfavourable to seedling establishment. Thus, actual gene flow may be recruitment limited rather than dispersal limited (Imbert and Lefèvre, 2003). In fact, comparison of total gene flow estimated here from non-dispersed seeds with total gene flow estimated from newly established seedlings in three F. excelsior remnants shows that actual recruitment of genes carried by immigrant pollen is limited (Table 4). Note that the ratio of potential to realized pollen-mediated gene flow is low, not because there seems to be an advantage conferred to recruitment of local seeds fertilized with local pollen but because the majority of establishing seedlings have immigrated into the remnants (Bacles et al., 2006). This indicates that in cases when seed dispersal is an important vector of long-distance dispersal estimating seed-mediated gene flow is essential to predicting landscape connectivity (Sork and Smouse, 2006).
In the southern uplands of Scotland and in other severely deforested landscapes, conservation management aimed at sustainable forest restoration without human intervention must move away from conservation gardening (Hobbs, 2007). Predictive conservation necessitates better understanding of the evolution of dispersal in a changing environment (Kokko and Lopez-Sepulcre, 2006) and an appreciation of how population genetic processes operate in ecological space and time. Bringing knowledge of contemporary gene flow among population remnants generated from this study and others into conservation will ensure that the evolutionary processes maintaining genetic connectivity and evolutionary potential are restored at the landscape scale (Meagher, 2007).
References
Aldrich PR, Hamrick JL (1998). Reproductive dominance of pasture trees in a fragmented tropical forest mosaic. Science 28: 103–105.
Amos W, Hoffman JI, Frodsham A, Zhang L, Best S, Hill AVS et al. (2007). Automated binning of microsatellite alleles: problems and solutions. Mol Ecol Notes 7: 10–14.
Araki H, Blouin MS (2005). Unbiased estimation of relative reproductive success of different groups: evaluation and correction of bias caused by parentage assignment errors. Mol Ecol 14: 4097–4109.
Bacles CFE, Burczyk J, Lowe AJ, Ennos RA (2005). Historical and contemporary mating patterns in remnant populations of the forest tree Fraxinus excelsior L. Evolution 59: 979–990.
Bacles CFE, Lowe AJ, Ennos RA (2006). Effective seed dispersal across a fragmented landscape. Science 311: 628.
Bittencourt JVM, Sebbenn AM (2007). Patterns of pollen and seed dispersal in a small fragmented population of the wind-pollinated tree Araucaria angustifolia in southern Brazil. Heredity 99: 580–591.
Brachet S, Jubier MF, Richard M, Jung-Muller B, Frascaria-Lacoste N (1999). Rapid identification of microsatellite loci using 5′ anchored PCR in the common ash Fraxinus excelsior. Mol Ecol 8: 157–168.
Burczyk J, Adams WT, Moran GF, Griffin AR (2002). Complex patterns of mating revealed in a Eucalyptus regnans seed orchard using allozyme markers and the neighbourhood model. Mol Ecol 11: 2379–2391.
Burczyk J, Chybicki IJ (2004). Cautions on direct gene flow estimation in plant populations. Evolution 58: 956–963.
Dakin EE, Avise JC (2004). Microsatellite null alleles in parentage analysis. Heredity 93: 504–509.
Dick CW (2001). Genetic rescue of remnant tropical trees by an alien pollinator. Proc Roy Soc Lond B 268: 2391–2396.
Dow BD, Ashley MV (1998). High levels of gene flow in bur oak revealed by paternity analysis using microsatellites. J Hered 89: 62–70.
Ferrazzini D, Monteleone I, Belletti P (2007). Genetic variability and divergence among Italian populations of common ash (Fraxinus excelsior L.). Ann For Sci 64: 159–168.
FRAXIGEN (2005). Ash Species in Europe: Biological Characteristics and Practical Guidelines for Sustainable Use. Oxford Forestry Institute, University of Oxford: UK, pp 128.
Gerard PR, Klein E, Austerlitz F, Fernandez-Manjarres JF, Frascaria-Lacoste N (2006). Assortative mating and differential male mating success in an ash hybrid zone population. BMC Evol Biol 6: 96.
Gerber S, Chabrier P, Kremer A (2003). FAMOZ: a software for parentage analysis using dominant, codominant and uniparentally inherited markers. Mol Ecol Notes 3: 479–481.
Gerber S, Mariette S, Streiff R, Bodenes C, Kremer A (2000). Comparison of microsatellites and amplified fragment length polymorphism markers for parentage analysis. Mol Ecol 9: 1037–1048.
Hadfield JD, Richardson DS, Burke T (2006). Towards unbiased parentage assignment: combining genetic, behavioural and spatial data in a Bayesian framework. Mol Ecol 15: 3715–3730.
Hamrick J (2004). Response of forest trees to global environmental changes. For Ecol Manage 197: 323–335.
Hazlitt SL, Sigg DP, Eldridge MDB, Goldizen AW (2006). Restricted mating dispersal and strong breeding group structure in a mid-sized marsupial mammal (Petrogale penicillata). Mol Ecol 15: 2997–3007.
Hebel I, Aldinger E, Haas R, Karopka M, Bogenrieder A, Dounavi A et al. (2007). Pollen dispersal and pollen contamination in an ash seed orchard (Fraxinus excelsior L.). Allgemeine forst und jagdzeitung 178: 44–49.
Heuertz M, Hausman F, Hardy OJ, Vendramin GG, Frascaria-Lacoste N, Vekemans X (2004). Nuclear microsatellites reveal contrasting patterns of genetic structure between western and southeastern European populations of the common ash (Fraxinus excelsior L.). Evolution 58: 976–988.
Heuertz M, Hausman F, Tsvetkov I, Frascaria-Lacoste N, Vekemans X (2001). Assessment of genetic structure within and among Bulgarian populations of the common ash (Fraxinus excelsior L.). Mol Ecol 10: 1615–1623.
Hobbs R (2007). Managing plant populations in fragmented landscapes: restoration or gardening? Aus J Bot 55: 371–374.
Hoffman JI, Amos W (2005). Microsatellite genotyping errors: detection approaches, common sources and consequences for paternity exclusion. Mol Ecol 14: 599–612.
Imbert E, Lefèvre F (2003). Dispersal and gene flow of Populus nigra (Salicaceae) along a dynamic river system. J Ecol 91: 447–456.
Janzen DH (1986). Blurry catastrophes. Oikos 47: 1–2.
Jones AG, Ardren WR (2003). Methods of parentage analyses in natural populations. Mol Ecol 12: 2511–2523.
Julien M-P, Alard D, Balent G (2006). Patterns of ash (Fraxinus excelsior L.) colonization in mountain grasslands: the importance of management practices. Plant Ecol 183: 177–189.
Jump AS, Penuelas J (2006). Genetic effects of chronic habitat fragmentation in a wind-pollinated tree. Proc Natl Acad Sci USA 103: 8096–8100.
Klein E, Lavigne C, Gouyon P-H (2006). Mixing of propagules from discrete sources at long distances: comparing a dispersal tail to an exponential. BMC Ecol 6: 3.
Kokko H, Lopez-Sepulcre A (2006). From individual dispersal to species range: perspectives in a changing world. Science 313: 789–790.
Lefort F, Brachet S, Frascaria-Lacoste N, Edwards KJ, Douglas GC (1999). Identification and characterization of microsatellite loci in ash (Fraxinus excelsior L.) and their conservation in the olive family (Oleaceae). Mol Ecol 8: 1075–1092.
Lowe AJ, Boshier DH, Ward M, Bacles CFE, Navarro C (2005). Genetic resource impacts of habitat loss and degradation: reconciling empirical evidence and predicted theory for neotropical trees. Heredity 95: 255–273.
Marshall TC, Slate J, Kruuk LEB, Pemberton J (1998). Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol 7: 639–655.
Meagher TR (2007). Paternity analysis in a fragmented landscape. Heredity 99: 563–564.
Morand ME, Brachet S, Rossignol P, Dufour J, Frascaria-Lacoste N (2002). A generalized heterozygote deficiency assessed with microsatellites in French common ash populations. Mol Ecol 11: 377–385.
Morissey MB, Wilson AJ (2005). The potential costs of accounting for genotypic errors in molecular parentage analyses. Mol Ecol 14: 4111–4121.
Nathan R (2006). Long-distance dispersal of plants. Science 313: 786–788.
Nielsen R, Mattila DK, Clapham PJ, Palsbøll PJ (2001). Statistical approaches to paternity analysis in natural populations and applications to the North Atlantic Humpback whale. Genetics 157: 1673–1682.
O′Connell LM, Mosseler A, Rajora OP (2006). Impacts of forest fragmentation on the mating system and genetic diversity of white spruce (Picea glauca) at the landscape scale. Heredity 97: 418–426.
Oddou-Muratorio S, Houot M-L, Demesure-Musch B, Austerlitz F (2003). Pollen flow in the wild service tree, Sorbus torminalis (L.) Crantz I. Evaluating the paternity analysis procedure in continuous populations. Mol Ecol 12: 3427–3439.
Ritland K (2002). Extensions of models for the estimation of mating systems using n independent loci. Heredity 88: 221–228.
Signorovitch J, Nielsen R (2002). PATRI—paternity inference using genetic data. Bioinformatics 18: 341–342.
Slate J, Marshall T, Pemberton J (2000). A retrospective assessment of the accuracy of the paternity inference program CERVUS. Mol Ecol 9: 801–808.
Slavov GT, Howe GT, Gyaourova AV, Birkes DS, Adams WT (2005). Estimating pollen flow using SSR markers and paternity exclusion: accounting for mistyping. Mol Ecol 14: 3109–3121.
Smouse PE, Sork VL (2004). Measuring pollen flow in forest trees. For Ecol Manage 197: 21–38.
Sork VL, Davis FW, Smouse PE, Apsit PE, Dyer RJ, Fernandez JF et al. (2002). Pollen movement in declining populations of California Valley Oak, Quercus lobata: where have all the fathers gone? Mol Ecol 11: 1657–1668.
Sork VL, Nason JD, Campbell DR, Fernandez JF (1999). Landscape approaches to historical and contemporary gene flow in plants. Trends Ecol Evol 14: 219–224.
Sork VL, Smouse PE (2006). Genetic analysis of landscape connectivity in tree populations. Landsc Ecol 21: 821–836.
Wagner AP, Creel S, Kalinowski ST (2006). Estimating relatedness and relationships using microsatellite loci with null alleles. Heredity 97: 336–345.
White GM, Boshier DH, Powell W (2002). Increased pollen flow counteracts fragmentation in a tropical dry forest: an example from Swietenia humilis Zuccarini. Proc Natl Acad Sci USA 99: 2038–2042.
Whitlock MC, McCauley DE (1999). Indirect measures of gene flow and migration: Fst≠1/(4Nm+1). Heredity 82: 117–125.
Young A, Boyle T, Brown AHD (1996). The population genetic consequences of habitat fragmentation for plants. Trends Ecol Evol 11: 413–418.
Acknowledgements
This study was carried out between 2000 and 2003 with financial support from The Scottish Forestry Trust (registered charity no. SC008465) to RAE in partial fulfilment of CFEB's doctorate of philosophy of the University of Edinburgh which was also supported by a PhD studentship of the Natural Environment Research Council. CFEB also acknowledges current funding from an outgoing international fellowship of the European Union (Marie Curie Individual Actions, contract CT-2005-9114 ADaPtE) for writing this article. We thank, first and foremost, AJ Lowe for use of sequencing facilities at CEH Edinburgh and advice to CFEB during the completion of her PhD. We thank S Cavers, J Cottrell and R Munro, for advice in the laboratory. Thank you to AC Newton and The Wildwood Group of the Borders Forest Trust. We also thank Tilhill Forestry, J Barker and C Moffat for field access in Moffat Dale, especially during the foot-and-mouth outbreak and M Young for help with sample collection. S Gerber and E Klein provided useful advice on gene flow inference in FAMOZ. We are indebted to J Pemberton and R Nathan and three anonymous reviewers for their insightful comments that contributed to improving this article.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary information
Rights and permissions
About this article
Cite this article
Bacles, C., Ennos, R. Paternity analysis of pollen-mediated gene flow for Fraxinus excelsior L. in a chronically fragmented landscape. Heredity 101, 368–380 (2008). https://doi.org/10.1038/hdy.2008.66
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2008.66
Keywords
This article is cited by
-
Pollination success of Fraxinus excelsior L. in the context of ash dieback
Annals of Forest Science (2023)
-
Characterising the molecular diversity of ash (Fraxinus excelsior L.) at its western marginal range in Europe — phylogeographic insights and implications for conservation in Ireland
Tree Genetics & Genomes (2022)
-
Gene flow and reproductive success in ash (Fraxinus excelsior L.) in the face of ash dieback: restoration and conservation
Annals of Forest Science (2021)
-
Wind pollination over 70 years reduces the negative genetic effects of severe forest fragmentation in the tropical oak Quercus bambusifolia
Heredity (2020)
-
Distinguishing mutations and null alleles from genotyping errors using mother progeny comparisons in Brazilian pine (Araucaria angustifolia)
Tree Genetics & Genomes (2019)