Abstract
We found significant population structure and isolation by distance among samples of flounder (Platichthys flesus) in the Baltic, Kattegat and Skagerrak seas using microsatellite genetic markers. This pattern was almost entirely due to a difference between flounder that have demersal spawning in the northern Baltic, as compared to pelagic spawners in the southern Baltic and on the west coast of Sweden. Among demersal spawners we found neither genetic differentiation nor any isolation by distance among sampling sites. We speculate that demersal flounder are descendants of a population that colonized the Baltic previous to pelagic spawners. The demersal flounder may thus have had longer time to adapt to the low salinity in the Baltic, and accordingly display egg characteristics that make it possible to reproduce at the low salinity levels in the northern Baltic. Among pelagic spawners significant isolation by distance was detected. Pelagic spawners have previously been shown to display clinal variation in egg size, which allows them to float also at the moderate salinity levels up to the region north of the island Bornholm. Management units for harvesting should ideally be based on true biological populations, and for the commercially important flounder up to 15 different management stocks in the Baltic have been suggested. We could not find a population genetic foundation for such a high number of management units, and our data suggest three management units: the northern Baltic (demersal populations), southern Baltic with the Öresund straits and the most northwestern sampling sites (Skagerrak, Kattegat and North Sea).
Similar content being viewed by others
Introduction
The Baltic Sea, being the largest brackish water area in the world, offers a challenge for both marine and freshwater species to live and prosper in (Voipio, 1981; Ojaveer and Lehtonen, 2001; Johannesson and André, 2006). The main limitation for marine species in the Baltic is the decreasing salinity with distance from the Öresund straits, whereas for freshwater species the problem is reversed. This is the main reason why the number of species is so low in the Baltic compared to adjacent areas. For marine fish, less than half of the approximately 120 species present in the North Sea thrive in the central Baltic Sea and in the Gulf of Bothnia and Gulf of Finland less than 20 marine fish species occur regularly (Voipio, 1981). Low salinity can be a problem for successful reproduction in marine teleosts. Low salinity immobilizes sperm, rendering many of the eggs unfertilized, and also diminishes egg survival (Holliday, 1969; Nissling et al., 2002, 2006). Another problem with the low salinity is the reduced buoyancy of eggs leading to pelagic eggs sinking into the more oxygen-depleted water where successful development is not possible. Nevertheless, many marine species, including fishes, have adapted to the low salinity in the Baltic. Several species have solved the problem with buoyancy by development of larger eggs, with less density (Mielck and Künne, 1932; Lönning and Solemdal, 1979; Thorsen et al., 1996; Nissling and Westin, 1997; Nissling et al., 2002). Recent studies have suggested that some of these adaptations may have resulted in genetic differentiation between the Baltic populations of a species and its marine counterpart (Nielsen et al., 2003, 2004; Jørgensen et al., 2005; Johannesson and André, 2006; but see Florin and Höglund, 2007).
Fish stocks are today mainly managed in traditional, political and practical management units. It is often not known, however, whether these management units also reflect biological stock units, that is if the harvest stock also represents a true population in the biological sense, with discrete population dynamics, or even an evolutionary significant unit (Begon et al., 1990; Carvalho and Hauser, 1994; Fraser and Bernatchez, 2001). The management of fish stocks would be more effective if it were actually based on true biological stocks rather than arbitrary defined stocks (Carvalho and Hauser, 1994), and there could be severe effects of lumping together populations that have separate population genetics and population dynamics when managing species (Ryman et al., 1995; Bailey, 1997; Frank and Brickman, 2001; Laikre et al., 2005). In the northeast Atlantic, assessment and management are often made according to the subdivisions (SD) determined by the International Council for the Exploration of the Sea (ICES) (Figure 1).
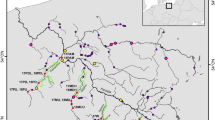
Map of sampling locations, International Council for the Exploration of the Sea (ICES) subdivisions (22–32, salinity and water depth (data from Seifert et al., 2001). (1) Finbo, (2) Åbo, (3) Gotska Sandön, (4) Hiiumaa, (5) Kvädöfjärden, (6) Muuga, (7) Helsinki, (8) Irbe, (9) Gotland, (10) Latvian Sea, (11) Hoburgsbank, (12) Smiltyne, (13) Bornholm, (14) Gdynia, (15) Dabki, (16) Oderbank, (17) Barsebäck, (18) Kungsbackafjorden, (19) Gullmaren and (20) Thyborön. Circles correspond to putative pelagic whereas squares represent putative demersal samples. Filled and open circles correspond to the suggested management units in the discussion.
The European flounder (Platichthys flesus) is the most commercially important flatfish in the Baltic Sea with yearly international landings of 15 000 tonnes. The flounder is assumed to be divided into several local populations, but few genetic or ecological studies have confirmed this idea. The flounder is distributed from the Skagerrak far up into the Baltic Sea. It is less frequently observed north of the Sea of Åland, and rarely north of the Northern Quark (Curry-Lindahl, 1985; Bagge and Steffensen, 1989; Muus et al., 1999; Voigt, 2002; Florin, 2005). Flounder migrate into less saline waters, and closer to the shore in shallower water, than other flatfish (Molander, 1964; Voigt, 2002; Florin, 2005).
The flounder could thus be considered both a coastal species and a migratory species. In general, flounder feed in shallow waters and migrate to spawn in deep waters; however, in the central and northern Baltic, flounder both spawn and feed in shallow water (Ehrenbaum, 1909; Molander, 1923, 1925, 1964; Aro, 1989; Florin, 2005). Tagging of adult flounders has revealed that migration in the southern Baltic is not so substantial, hence it is highly plausible that the flounder could be divided into local populations (Otterlind, 1967). The same was shown in the central Baltic (Otterlind, 1966), in this case however, single individuals did undertake substantial movement (hundreds of km).
On the basis of tagging, several harvest stocks and potential biological populations of flounder have been identified in the Baltic. In the management units SD 22 and 23 (Figure 1) three and one local stocks were identified, respectively (Bagge, 1966; Bagge and Steffensen, 1989). Further tagging studies in SD 24 and 25 indicated that each region supported a distinct stock (Otterlind, 1967). Tagging experiments in SD 26 and 28 (Cieglewicz, 1947, 1961, 1963; Otterlind, 1967; Vitins̆, 1972; Bagge and Steffensen, 1989) suggested that there were two stocks in each SD. The Gotland basin, with low oxygen content, seems to prevent flounder from crossing over and may act as an east–west barrier (Aro, 1989; Bagge and Steffensen, 1989). It is unclear if SD 27 supports one (Aro, 1989) or two (Bagge and Steffensen, 1989) stocks of flounder. Tagging experiments in SD 29, 30 and 32 (reviewed in Aro, 1989) suggested that there was one stock of flounder in SD 29 and 30 and a separate stock in SD 32. Ojaveer et al. (1985) further speculated that flounders in SD 32 are divided into two stocks—one along the Finnish coast and one along the coast of Estonia. This gives in total 15 potential stocks of flounder in the Baltic Sea. It remains, however, to be discerned whether these are true biological, genetically different, stocks or ‘merely’ harvest stocks.
The development of larger eggs, with lower specific gravity, as an adaptation to less saline water is seen in flounder (Mielck and Künne, 1932; Lönning and Solemdal, 1979; Nissling et al., 2002). Although some of the response may be due to plasticity, transplantation experiments suggest that flounders, like cod, have distinct populations with distinct innate egg properties, and a limited ability to acclimatize to new salinities (Solemdal, 1967, 1971, 1973; Thorsen et al., 1996; Nissling and Westin, 1997; Nissling et al., 2002). The maximum size of flounder eggs (that is those with minimum specific gravity) is found in waters of 10–12‰ salinity. This may indicate that eggs cannot be buoyant in water of lower salinity. Supporting this conclusion, Mielck (1926) found no floating flounder eggs above 40 m depth to the north and west of Bornholm, nor above 50 m in the deep of the Bornholm Basin nor above 100 m in the deep area in the Gdansk basin and east of Gotland. This corresponds to a 10–11‰ isohaline. For flounder in SD 24 and 25 the appropriate habitat for successful reproduction has a minimum salinity of approximately 12‰ and minimum oxygen concentration of 2 ml l−1 (ACFM, 2005). This means that the recruitment success fluctuates depending on the hydrological conditions on the spawning ground. According to calculations by Nissling et al. (2002), successful reproduction for pelagic flounder may occur regularly in the Sound, the Arkona and the Bornholm basins and, during favourable conditions, in the Gdansk and Gotland basins. The same authors also found a significant difference in the salinity required for neutral egg buoyance for flounders collected from the Sound, compared to those from the Arcona or Bornholm basin. There was also a significant increase in mean egg size from 1.12 mm in the Sound to 1.34 mm in the Arcona basin and finally to 1.43 mm in the Bornholm basin (Nissling et al., 2002).
Reproductive populations of flounders do, however, also exist on the shallow central banks and in the eastern part of the Baltic with water of only 5–7‰ salinity. Eggs from these areas are smaller and heavier and develop at the bottom (Sandman, 1906; Mielck, 1926; Mielck and Künne, 1932; Solemdal, 1967, 1971; Lönning and Solemdal, 1979; Bonsdorff and Norkko, 1994; Nissling et al., 2002). According to Nissling et al. (2002) the mean egg size in the Eastern Gotland basin was only 0.99 mm. Presumably, selection has favoured tougher, heavier eggs that are better at resisting the mechanical forces acting at the bottom (Solemdal, 1967, 1971). The existence of two separate reproductive patterns in the Baltic is further supported by Mielck (1926) and Mielck and Künne (1932) who caught ripe female species at Oderbank and Mittelbank, locations with 6–7‰ salinity. Some of the female species had normal, small ‘bank’ type of eggs, but also a few were found with large ‘pelagic’ eggs. It is uncertain if individual flounders can change spawning behaviour and type of eggs between years, or if there are truly two different, genetically distinct stocks of flounder. The demersal spawning flounder, presumably constituting one distinct stock with respect to salinity requirements for reproduction may spawn successfully as far north as the southern Gulf of Bothnia and the Gulf of Finland (Nissling et al., 2002).
In this study we used microsatellite DNA to map the genetic population structure of flounder in the Baltic and in the Skagerrak/Kattegat area. We were in particular interested to test the hypothesis that flounder form different populations, and if so whether they correspond to demersal and pelagic flounder and whether the clinal variation in egg characteristics and spawning behaviour correspond with genetical differences at neutral microsatellite loci.
Materials and methods
Sampling
In spring and early summer 2003, tissue samples from the base of the pectoral fin were taken from approximately 50 adult fish from 20 localities. Effort was made to sample as close to spawning time as possible. Flounder spawn between February and April in the North Sea, Skagerrak and Kattegat. In the Baltic spawning is delayed eastwards and northwards, so that around Gotland spawning takes place in April–June and in the Gulf of Finland in May–June (Molander, 1964; Curry-Lindahl, 1985). For practical and jurisdictional reasons, however, some locations were sampled prior to or just after the spawning period. The different sampled areas ranged from the Åland Sea to the North Sea, and were Finbo, Åbo, Gotska Sandön, Hiiumaa, Kvädöfjärden, Muuga, Helsinki, Irbe, Gotland, Latvian Sea, Hoburgsbank, Smiltyne, Bornholm, Gdynia, Dabki, Oderbank, Barsebäck, Kungsbackafjorden, Gullmaren and Thyborön. Sample statistics and locations are shown in Table 1 and Figure 1, respectively.
Molecular methods
DNA was extracted using 500 μl 5% Chelex 100 resin (Bio-Rad Laboratories, Hercules, CA, USA) solution with 4 μl 10 mg ml−1 proteinase K. Samples were incubated at 56 °C on a rocking platform for 2 h, vortexed for 15 s, heated to 96 °C for 15 min in temperature blocks and finally vortexed again for 15 s. Samples were centrifuged at 13 000 r.p.m. for 5 min and the supernatant was taken for use in PCR.
Seven microsatellite loci (Table 2, sequences obtained from GenBank, submitted by TJ Dixon 2001) were typed for all individuals. The loci AJ315971 and AJ315975 were amplified together in a multiplex PCR and the same was true for AJ315970 and AJ315974. PCR reactions (10 μl) were set up in 96-well PCR plates using the final concentrations: 1 × NH4 reaction buffer (BioLine, London, UK), 1 mM MgCl2, 0.13 mM of each of the four deoxyribonucleotide triphosphates, 1 μM of the reversed primer, 0.5 μM labelled forward primer, 0.5 μM unlabelled forward primer 0.25 units Taq polymerase (BioLine, Biotaq, London, UK) and 1 μl DNA (10–100 ng). The amplifications were run on an Eppendorf Mastercycler Gradient or a Geneamp PCR system 2700 (Applied Biosystems, Foster City, CA, USA) using the following temperature cycle: initial denaturing at 96 °C for 3 min followed by 32 cycles of 50 s at 95 °C, 50 s Ta °C and 70 sat 72 °C and finally there was a 4 °C hold. Annealing temperatures (Ta) are given in Table 2.
Fragment sizes were analysed using an ABI Prism 377 DNA sequencer according to the manufacturers’ protocol. Two to five different loci were run together in a single line on the sequencer. A mix of samples was made by taking 2 μl of each PCR reaction and diluting with 5 μl dd H2O. This mix (1 μl) was loaded onto the gel together with 0.29 μl TAMRA 500 size standard and 0.71 μl formamid loading dye. Gel images were analysed using the ABI Prism 377 DNA sequencer data collection software GeneScan 2.1.1.
Statistical analyses
Observed and Nei's unbiased expected heterozygosity were estimated for each population and locus using FSTAT 2.9.3 (Goudet, 1995, 2001). Using the same software, deviations from Hardy–Weinberg (HW) equilibrium were tested by 2800 permutations and the independence of loci was tested with G-statistics and 8400 permutations. Both global and pair-wise FST (Weir and Cockerham, 1984) were calculated using FSTAT 2.9.3 that also provided confidence interval for the global FST using bootstrapping over loci (15 000 replacements) and significance testing of pair-wise FST:s by G-statistics and 3800 permutations. FSTAT 2.9.3 was further used to analyse isolation by distance (Mantel tests with 10 000 permutations) and perform partial Mantel tests (10 000 randomizations). GENEPOP 3.4 (Raymond and Rousset, 1995) was used to calculate P-values for single loci FST that were combined according to Fisher's method (Sokal and Rohlf, 1995) to provide a P-value for the global FST. GENETIX 4.05 (Belkhir, 2004) was used to calculate pair-wise genetic distances sensu (Nei, 1972). The latter was further analysed using multidimensional scaling (MDS) with an alternating least-squares algorithm in SPSS 12.0 (SPSS Inc.) and a Young's S-stress convergence limit of 0.001. Tests for the presence of null alleles and scoring errors were made with MICRO-CHECKER 2.2 (Van Oosterhout et al., 2004).
Geographical statistics and calculations of distance between sampling areas were made with ARCGIS 9.1 (ESRI). Inference of number of populations and locations of possible genetic boundaries without assuming any a priori population structure was made using the R package GENELAND (Guillot et al, 2005b). By implementation of a Bayesian cluster model into a Markov Chain Monte Carlo algorithm each individual was probabilistically assigned to a population (Pritchard et al., 2000). We used 100 000 iterations, an uncertainty of coordinates of 1 km, and possible number of populations between 1 and 20. The model was re-run with 50 000 iterations, a burn-in period of 50 iterations and the estimated number of populations from the first run fixed. Voronoi tessellation of observed genetic data resulted in maps of the posterior probability of belonging to a certain population (see Guillot et al, 2005a,2005b for a more extensive description of the technique).
Environmental information about the sampling locations, that is temperature and salinity, was extracted from the oceanographic surface database of ICES, the oceanographic database of the Swedish Meteorological and Hydrological Institute and the monitory fishing database of the Institute of Coastal Research at the Swedish Board of Fisheries. Data were restricted to less than 10 m depth and the summertime (May–August) during the time period 1993–2003. Observations within a 30-km radius from the sampling location and mean values for temperature and salinity were used.
Results
To test for laboratory artefacts one sample (Oderbank) was run twice. We obtained very similar results, with 95% of allele sizes being the same, and the same qualitative conclusions regardless of run used. Total allele numbers ranged between 8 and 41 per locus and between 3 and 34 within samples. The mean observed heterozygosity within samples varied between 0.65 and 0.83, whereas the observed heterozygosity per locus per sample varied between 0.50 and 0.97 (see appendix for details). Tests for linkage disequilibrium were nonsignificant after Bonferroni correction but two samples (Åbo and Gdynia) showed deviations from HW expectations at the AJ315972 locus. Furthermore, MICRO-CHECKER suggested that there could be a problem with null alleles at some loci in some populations, although the estimated frequencies of null alleles were always <0.1. Therefore, in addition to using the original data, all statistical analyses were also made excluding the most problematic locus (AJ315972), which had an estimated mean null allele frequency above 0.05 in six of the samples. However, including or excluding this locus did not qualitative change the outcome of these tests and hence only results from using all loci are presented.
Genetic differentiation
The global FST (Weir and Cockerham, 1984) including all samples was 0.012 (Fisher's method of test combination, χ2=152, d.f.=14, P<0.001; 95% CI 0.008–0.016). Total 58 of 190 pair-wise estimates of FST were significant after Bonferroni correction. Ten of these involved either one of the most separate samples; Finbo or Thyborön (Table 3). The highest pair-wise FST was 0.048 between the Gotland and the Dabki samples.
Using only the samples from the Baltic Sea (including Öresund but excluding Kattegat) resulted in a global FST of 0.011 (95% CI 0.007–0.016). The samples later identified as demersal (see below: 1–9, 11 and 12) had a nonsignificant global FST although there was a global FST of 0.006 (95% CI 0.003–0.011) among the putative pelagic samples.
Isolation by distance
There was a significant correlation between nearest waterway distance (in km) and genetic distance (FST/(1−FST)) (Mantel test, R2=0.23, P=0.0001). A partial mantel test, with the ecological indicator variables salinity and temperature together with geographical distance, showed that only geographic distance was correlated to genetic distance.
A plot of genetic distance and geographic distance showed that samples 2–9, 11 and 12 were more alike the Finbo (1) sample than the rest (Figure 2a). The most striking deviation from the genetic–geographic distance relationship was shown by sample 10 (Latvian Sea). If samples were divided in potential demersal and pelagic stocks, according to the MDS plot (see below), it was revealed that the isolation by distance pattern was driven by the pelagic spawners (R2=0.25, P=0.002), but not the demersal (R2=0.007, P=0.54; Figure 2b).
(a) Relationship between genetic distance (FST) and geographic distance from the northernmost sample at Åland. (b) Relationship between linearized genetic distance (FST/(1−FST)) and the logarithm of geographic distance (filled squares, putative demersal spawners; open circles, putative pelagic spawners).
Number of populations
The most likely number of populations was two (GENELAND; Figure 3a). A map of posterior probabilities showed that the northern Baltic samples were more likely to belong to one population, whereas the southern Baltic samples and the Kattegat, Skagerrak and North Sea samples belonged to another (Figure 3b).
(a) Posterior distribution of the estimated number of populations in 100 000 MCMC iterations in GENELAND. (b) Map of posterior probabilities of population membership (number of populations=2 and 50 000 iterations). Lighter areas correspond to higher probability to belong to the demersal population, sampling locations are indicated with white dots.
Multidimensional scaling
An MDS analysis of pair-wise Nei's genetic distances (Nei, 1972) showed that the first dimension explained 81% of the variance (one-dimensional r2=0.81). The second dimension explained an additional 8% (two-dimensional r2=0.89) and the third 4% (three-dimensional r2=0.93). There was no clear geographic trend correlated to dimension 1, but a general east/west division could be detected (Figure 4a), and the North Sea, Skagerrak and Kattegat samples were clustered together.
(a) Multidimensional scaling (MDS) plot of Nei's distance among samples, one-dimensional r2=0.81, two-dimensional r2=0.89. (b) MDS plot among potential demersal samples, one-dimensional r2=0.28, two-dimensional r2=0.54. (c) MDS plot of Nei's genetic distances among pelagic spawners, one-dimensional r2=0.37; two-dimensional r2=0.81.
An MDS plot of potential demersal samples showed that there was no pattern among these (Figure 4b). However, among the pelagic samples the Skagerrak, Kattegat and North Sea samples were clustered together (Figure 4c)
Discussion
In accordance with many population genetic studies of organisms inhabiting both the North Sea and the Baltic (reviewed in Johannesson and André, 2006), the flounder show clinal variation in allele frequencies from the west coast of Sweden to the northern areas of the Baltic. However, not all studies have revealed this pattern. In a study of turbot (Psetta maxima, Florin and Höglund, 2007) we found no evidence of isolation by distance or population structure, despite the fact that previous works (Nielsen et al., 2004) have suggested differences between the North Sea and the Baltic for this species. We attributed the absence of population structure in our study to admixture during major salt-water influxes in to the Baltic; events that occur stochastically with approximately 10-year intervals (Fonselius and Valderrama, 2003).
We found a clear pattern of isolation by distance in the present study of the flounder, and a closer examination of the map of posterior probabilities of population membership, as well as the isolation by distance plots, reveals that the pattern to a large extent is generated by a stepped cline situated somewhere in the region of the island of Bornholm. This stepped cline coincides with a difference in spawning behaviour in the flounder. North of Bornholm, the flounder is largely resident and spawns at shallow waters with demersal eggs. These demersal eggs also show evidence of adaptations to lower salinity by having a thicker chorion and being more robust (Lönning and Solemdal, 1979). South of Bornholm, the flounder is migratory and spawns in deeper waters with pelagic eggs.
The evidence for the existence of a genetically distinct demersal type of flounder is indirect, but fits with our observation of a stepped cline in allele frequencies, as well as the map of posterior probabilities of population memberships. Although tagging experiments (reviewed in Aro and Sjöblom, 1983; Florin, 2005) have revealed extensive migratory behaviour of flounder there are no direct estimates of migration between demersal and pelagic populations. Extensive gene flow between populations in the Baltic would counteract any local adaptations to salinity. However, stepped clines can evolve despite gene flow, provided that local selection pressures are strong enough (Endler, 1977, but see Vasemägi, 2006).
Within samples taken from demersal populations we could find no evidence of population structure, suggesting that gene flow is extensive among these populations. However, among pelagic samples, we found both a significant isolation by distance pattern, and that the most northwestern samples (the North Sea, Skagerrak and Kattegat) were differentiated from the rest of the pelagic samples. These observations argue that there is indeed a true isolation by distance pattern within pelagic spawners. This is in agreement with the observation of significantly different egg characteristics that has been demonstrated between flounders from the Sound and the southern Baltic Sea (Nissling et al., 2002). We speculate that the difference between demersal and pelagic spawners may be of a more ancient origin than the population structure observed within pelagic spawners. Such a scenario may also explain why we observe population structure in flounder, but not in turbot (Florin and Höglund, 2007). In flounder, the demersal spawners may be descended from a population that colonized the Baltic previous to present pelagic flounder populations and any population of turbot. This demersal stock of flounder may thus have had longer time to adapt to the brackish salinity found in the northern Baltic. One such key adaptation may be their thicker shelled eggs, a feature that may be essential for survival at low salinity. No such clear differentiation in egg characteristics can be observed in turbot.
Management
When considering management units it seems clear that from a strict population genetic perspective, on the basis of neutral (or at least nearly neutral) microsatellite markers, there are no arguments for more than three management units. These are the northern Baltic (demersal populations); southern Baltic with the Öresund straits and the most northwestern sampling sites (Skagerrak, Kattegat and North Sea), respectively. There is no population genetic foundation for the many SDs currently implemented. Furthermore, our data reveal that some of the SDs harbour flounder of both types. Unfortunately, from a management point of view, no clear geographic boundary between the demersal and pelagic flounder can be given. Even if, during spawning, they divide into shallower and deeper areas, respectively, they probably mix in the feeding season. The distribution of the pelagic type is most probably highly affected by the changing salinity due to shifting hydrological conditions. However, decisions about management units should best be based on more than one type of genetic marker, and also other relevant biological criteria such as ecological differences between populations (Ruzzante et al., 2006). Although our data strongly argue for three management units, we argue for further studies using more markers and ecological data to further strengthen the conclusions from this study. Genetic studies could be based on mitochondrial markers that give information on historical population divergence between different colonization events of the Baltic (cf. Macoma Baltica, Luttikhuizen et al., 2003), and single nucleotide polymorphisms reflecting adaptive differences among populations. Ecological studies could be based on tagging, differences in spawning behaviour and egg characteristics.
References
ACFM (2005). Report of the ICES Advisory Committee on Fishery Management,June 2005, 8, ICES: Copenhagen.
Aro E (1989). A review of fish migration patterns in the Baltic. Rapp P-v Réun Cons int Explor Mer 190: 72–96.
Aro E, Sjöblom V (1983). The migration of flounder in the Northern Baltic sea. ICES CM J: 1–12.
Bagge O (1966). Tagging of flounder in the Western Baltic, the Belt Sea and the Sound in 1960–62. ICES CM D: 15.
Bagge O, Steffensen E (1989). Stock identification of demersal fish in the Baltic. Rapp P-v Réun Cons int Explor Mer 190: 3–16.
Bailey KM (1997). Structural dynamics and ecology of flatfish populations. J Sea Res 37: 269–280.
Begon M, Harper JL, Townsend CR (1990). Ecology, 2nd edn, Blackwell: Oxford.
Belkhir K (2004). GENETIX, logiciel sous Windows™ pour la génétique des populations.Laboratoire Génome, Populations, Interactions CNRS UMR 5000, Université de Montpellier II, Montpellier (France).
Bonsdorff E, Norkko A (1994). Flounder (Platichthys flesus) spawning in Finnish archipelago waters. Memo Soc Fauna Flora Fenn 70: 30–31.
Carvalho GR, Hauser L (1994). Molecular genetics and the stock concept in fisheries. Rev Fish Biol Fish 4: 326–350.
Cieglewicz W (1947). The migration and the growth of the marked flounder from the Gulf of Gdansk and the Bornholm Basin. Archiwum hydrobiologii i Rybactwa XIII: 105–164.
Cieglewicz W (1961). Tagging experiments with flatfish in the southern Baltic. ICES CM Baltic-Belt Seas Committee No. 95: 2.
Cieglewicz W (1963). Flounder migration and mortality rates in the southern Baltic. ICES CM Baltic-Belt Seas Committee No. 78: 7.
Curry-Lindahl K (1985). Våra fiskar. Havs- och sötvattensfiskar i Norden och övriga Europa. P.A. Norstedt & Söners förlag: Stockholm.
Ehrenbaum E (1909). Eier und Larven der im Winter laichenden Fische der Nordsee II Die Laichverhältnisse von Scholle und Flunder. Arb dt wiss Komm int Meeresforsch 12: 1–176.
Endler JA (1977). Geographic Variation, Speciation and Clines. Princeton University Press: Princeton, NJ.
Florin A-B (2005). Flatfishes in the Baltic sea—a review of biology and fishery with a focus on swedish conditions. Finfo 2005: 14.
Florin A-B, Höglund J (2007). Abscence of population structure of turbot (Psetta maxima) in the Baltic Sea. Mol Ecol 16: 115–126.
Fonselius S, Valderrama J (2003). One hundred years of hydrographic measurements in the Baltic Sea. J Sea Res 49: 229–241.
Frank KT, Brickman D (2001). Contemporary management issues confronting fisheries science. J Sea Res 45: 173–187.
Fraser DJ, Bernatchez L (2001). Adaptive evolutionary conservation: towards a unified concept for defining conservation units. Mol Ecol 10: 2741–2752.
Goudet J (1995). FSTAT (vers. 1.2): a computer program to calculate F-statistics. J Hered 86: 485–486.
Goudet J (2001). FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Available fromhttp://www.unil.ch/izea/softwares/fstat.html.Updated from Goudet (1995).
Guillot G, Estoup A, Mortier F, Cosson JF (2005a). A spatial statistical model for landscape genetics. Genetics 170: 1261–1280.
Guillot G, Mortier F, Estoup A (2005b). Geneland: a computer package for landscape genetics. Mol Ecol Notes 5: 712–715.
Holliday FGT (1969). The effects of salinity on the eggs and larvae of teleosts.In: Hoar WS, Randall DJ (eds). Fish Physiology 1. Excretion, Ionic Regulation and Metabolism. Academic Press: New York. pp 293–311.
Johannesson K, André C (2006). Life on the margin: genetic isolation and diversity loss in a peripheral marine ecosystem, the Baltic Sea. Mol Ecol 15: 2013–2029.
Jørgensen HBH, Hansen MM, Bekkevold D, Ruzzante DE, Loeschke V (2005). Marine landscapes and population genetic structure of herring (Clupea harengus L.) in the Baltic Sea. Mol Ecol 14: 3219–3234.
Laikre L, Palm S, Ryman N (2005). Genetic population structure of fishes: implications for coastal zone management. Ambio 34: 111–119.
Lönning S, Solemdal P (1979). The relation between thickness of chorion and specific gravity of eggs from Norwegian and Baltic flatfish populations. FiskDir Skr Ser HavUnders 16: 77–88.
Luttikhuizen PC, Drent J, Baker AJ (2003). Disjunct distribution of highly diverged mitochondrial lineage clade and population subdivision in a marine bivalve with pelagic larval dispersal. Mol Ecol 12: 2215–2229.
Mielck W (1926). Untersuchungen über die pelagische Fischbrut (Eier und Larven) in der Ostsee im April 1925. Berichte der Deutsch. Wiss kommission f Meeresforschung 2: 311–318.
Mielck W, Künne C (1932). Fischbrut und Plankton—Untersuchungen auf dem Reichsforschungsdampfer ‘Poseidon’ in der Ostsee, mai-juni 1931. Wiss Meeresunters Abt Helgoland 19: 1–120.
Molander AR (1923). Undersökningar rörande flundran (Pleuronectes flesus L.) i Mellersta Östersjön. Medd Kungl Lantbruksstyrelsen 243: 1–24.
Molander AR (1925). Undersökningar över rödspotta (Pleuronectes platessa L.) flundra (Pleuronectes flesus L.) och sandskädda (Pleuronectes limanda L.). Svenska Hyd Biol Komm 1: 1–38.
Molander AR (1964). Underordning plattfiskar.In: Andersson KA (ed). Fiskar och fiske i norden. Natur och kultur: Stockholm.pp 90–113.
Muus BJ, Nielsen JG, Svedberg U (1999). Havsfisk och fiske i nordvästeuropa. Prisma: Stockholm.
Nei M (1972). Genetic distance between populations. Am Nat 106: 283–292.
Nielsen EE, Hansen MM, Ruzzante DE, Meldrup D, Grønkjær P (2003). Evidence of a hybrid-zone in Atlantic cod (Gadus morhua) in the Baltic and the Danish Belt Sea, revealed by individual admixture analysis. Mol Ecol 12: 1497–1508.
Nielsen EE, Nielsen PH, Meldrup D, Hansen MM (2004). Genetic population structure of turbot (Scophthalmus maximus L.) supports the presence of multiple hybrid zones for marine fishes in the transition zone between the Baltic Sea and the North Sea. Mol Ecol 13: 585–595.
Nissling A, Johansson U, Jacobsson M (2006). Effects of salinity and temperature conditions on the reproductive success of turbot (Scopthalmus maximus) in the Baltic Sea. Fish Res 80: 230–238.
Nissling A, Westin L (1997). Salinity requirements for successful spawning of Baltic and Belt Sea cod and the potential for cod stock interactions in the Baltic Sea. Mar Ecol Prog Ser 152: 261–271.
Nissling A, Westin L, Hjerne O (2002). Reproductive success in relation to salinity for three flatfish species, dab (Limanda limanda), plaice (Pleuronectes platessa) and flounder (Pleuronectes flesus), in the brackish water Baltic Sea. ICES J Mar Sci 59: 93–108.
Ojaveer E, Kaleis M, Aps R, Lablaika I, Vitins̆ M (1985). The impact of recent environmental changes on the main commercial fish stocks in the Gulf of Finland. Finnish Fish Res 6: 1–14.
Ojaveer E, Lehtonen H (2001). Fish stocks in the Baltic Sea: finite or infite resource. Ambio 30: 217–221.
Otterlind G (1966). Flundrans vandringsvanor i mellersta Östersjön. Ostkusten 38: 19–26.
Otterlind G (1967). Om rödspättans och flundrans vandringsvanor i södra Östersjön. Ostkusten 10: 9–14.
Pritchard JK, Stephens M, Donnelly P (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
Raymond M, Rousset F (1995). GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86: 248–249.
Ruzzante DE, Mariani S, Bekkevold D, André C, Mosegaard H, Clausen LAW et al. (2006). Biocomplexity in a highly migratory pelagic marine fish, Atlantic herring. Proc R Soc B 273: 1459–1464.
Ryman N, Utter F, Laikre L (1995). Protection of intraspecific biodiversity of exploited fishes. Rev Fish Biol Fish 5: 417–446.
Sandman VJA (1906). Kurzer bericht über in Finnland ausgeführte untersuchungen über den Flunder, den Steinbutt und den Kabeljau. Rapp P-v Réun Cons int Explor Mer 5: 37–44.
Seifert T, Tauber F, Kayser B (2001). A High Resolution Spherical Grid Topography of the Baltic Sea,2nd edn, Baltic Sea Science Congress: Stockholm.pp 25–29. November 2001, poster no. 147.
Sokal RR, Rohlf FJ (1995). Biometry,3d edn, WH Freeman: New York.
Solemdal P (1967). The effect of salinity on buoyancy, size and development of flounder eggs. Sarsia 29: 431–442.
Solemdal P (1971). Prespawning flounders transferred to different salinities and the effects on their eggs. Vie et Milieu Supplément 22: 409–423.
Solemdal P (1973). Transfer of Baltic flatfish to a marine environment and the long term effects on reproduction. Oikos 15: 268–276.
Thorsen A, Kjesbu OS, Fyhndr HJ, Solemdal P (1996). Physiological mechanisms of buoyancy in eggs from brackish water cod. J Fish Biol 48: 457–477.
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004). MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4: 535–538.
Vasemägi A (2006). The adaptive hypothesis of clinal variation revisited: single-locus clines as a result of spatially restricted gene flow. Genetics 173: 2411–2414.
Vitins̆ MJ (1972). Migration of flounder in the Eastern Baltic. ICES CM F: 12.
Weir BS, Cockerham CC (1984). Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370.
Voigt H-R (2002). Piggvaren i våra kustvatten. Fiskeritidskrift för Finland 1: 25–27.
Voipio A (ed). (1981). The Baltic Sea. Elsevier: Amsterdam.
Acknowledgements
The collection of genetic material would not have been possible without Kaj Ådjers at the Fisheries Division in the Provincial Government of Åland Islands, Claus-Christian Friees at the Institute for Baltic Sea Fisheries in Rostock, Didziz Ustup at Latvian Fish Resource Agency, Timo Myllelä and Eero Aro at the Finnish Game and Fisheries Research Institute, Tenno Drevs at the Estonian Marine Institute, Iwona Psuty-Lipska at the Sea Fisheries Institute in Gdynia, Jacob-Hemmer Hansen at the Danish Institute for Fisheries Research and the Fisheries Research Laboratory in Lithuania. Within Sweden the sampling was made with help of Anna Lingman, Fredrik Franzen, Mikael Petterson and Björn Fagerholm and Kurt Torildsson at the Institute of Coastal Research in Simpevarp and Ringhals, respectively and Anders Nissling at the Gotland University. We also want to thank Robert Ekblom and Gunilla Engström at the Department of Population Biology, Uppsala University for the molecular work and Carl André and one anonymous referee for valuable comments on the chapter.
Author information
Authors and Affiliations
Corresponding author
Appendix 1
Appendix 1
Summary of basic genetic data per sample and locus: number of scored individuals (N), number of alleles, allelic richness in a sample of 17 individuals (r(17)), observed (Hobs) and expected (Hexp) heterozygosity and P-values for deviation from Hardy–Weinberg equilibrium (HW). HW per locus per sample is based on 2800 randomizations. Adjusted 5% level is 0.00036.
Rights and permissions
About this article
Cite this article
Florin, AB., Höglund, J. Population structure of flounder (Platichthys flesus) in the Baltic Sea: differences among demersal and pelagic spawners. Heredity 101, 27–38 (2008). https://doi.org/10.1038/hdy.2008.22
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2008.22
Keywords
This article is cited by
-
Genetically Distinct European Flounder (Platichthys Flesus, L.) Matriline in the Black Sea
Thalassas: An International Journal of Marine Sciences (2024)
-
Ecological connectivity of the marine protected area network in the Baltic Sea, Kattegat and Skagerrak: Current knowledge and management needs
Ambio (2022)
-
Long-term changes in spatial overlap between interacting cod and flounder in the Baltic Sea
Hydrobiologia (2020)
-
Mind the gut: genomic insights to population divergence and gut microbial composition of two marine keystone species
Microbiome (2018)
-
Reconciling differences in natural tags to infer demographic and genetic connectivity in marine fish populations
Scientific Reports (2018)