Abstract
A variety of questions in population and evolutionary biology are studied using chloroplast DNA (cpDNA). The presumed maternal inheritance in angiosperms allows for certain assumptions and calculations to be made when studying plant hybridization, phylogeography, molecular systematics and seed dispersal. Further, the placement of transgenes in the chloroplast to lessen the probability of ‘escape’ to weedy relatives has been proposed since such genes would not move through pollen. In many studies, however, strict maternal inheritance is assumed but not tested directly, and some studies may have sample sizes too small to be able to detect rare paternal leakage. Here, we study the inheritance of cpDNA simple sequence repeats in 323 offspring derived from greenhouse crosses of the rare sunflower Helianthus verticillatus Small. We found evidence for rare chloroplast paternal leakage and heteroplasmy in 1.86% of the offspring. We address the question of whether one can extrapolate the mode of chloroplast transmission within a genus by comparing our results to the findings of another sunflower species study. The findings of occasional paternal transmission of the chloroplast genome are discussed in the framework of using these markers in studies of population and evolutionary biology both in Helianthus and other angiosperms.
Similar content being viewed by others
Introduction
Population and evolutionary biologists use chloroplast DNA (cpDNA) in a variety of applications, including studies of hybridization and phylogeography (Rieseberg and Ellstrand, 1993; Welch and Rieseberg, 2002; Dobes̆ et al., 2004; Van Droogenbroeck et al., 2006), plant molecular systematics (Olmstead and Palmer, 1994; Kelchner, 2000; Wolfe and Randle, 2004) and seed movement and dispersal in natural populations (McCauley, 1995; Ouborg et al., 1999; Hamilton and Miller, 2002; Petit et al., 2005). Since the chloroplast genome is largely maternally transmitted in angiosperms (Sears, 1980; Corriveau and Coleman, 1988; Zhang et al., 2003), these types of applications assume maternal inheritance (Birky, 1995, 2001), though this assumption is only rarely tested. In fact, occasional paternal or biparental inheritance has been shown in some species (Sears, 1980; Corriveau and Coleman, 1988; Reboud and Zeyl, 1994; Röhr et al., 1998; Hansen et al., 2007; McCauley et al., 2007). Thus, when it occurs, paternal or biparental inheritance of the chloroplast genome could lead to incorrect conclusions in studies involving seed dispersal, hybrid origins and evolutionary relationships should maternal inheritance be assumed.
Further, researchers have posited that transgenes placed in the chloroplast genome of crops would reduce their probability of ‘escape’ as the genes would not move through pollen if maternally inherited (Gressel, 1999; Daniell et al., 2005; Grevich and Daniell, 2005). In crop systems, non-maternal inheritance could lead to the escape of transgenes, for example that may confer herbicide resistance, thus leading to the possibility of creating ‘superweeds’ since many domesticated crops grow in close proximity to their weedy wild relatives (Smith, 1989; Haygood et al., 2004; Chapman and Burke, 2006). In fact, Haygood et al. (2004) found that even with low levels of paternal transmission, the probability of transgene escape is considerable. Therefore, studying the transmission of the chloroplast genome can provide valuable information for the likelihood of transgene escape in domesticated crop species as well as accepting or rejecting the assumptions of maternal inheritance in population and evolutionary studies.
In many plant genera, consistency of inheritance has been observed among the several congeners that happen to have been studied (Sears, 1980; Corriveau and Coleman, 1988; Zhang et al., 2003). These findings suggest that chloroplast inheritance may be conserved within a genus. If so, one could extend information on the mode of inheritance of one species to its congeners. However, there are a few exceptions to this observation in which different members of a genus have conflicting modes of inheritance (Sears, 1980; Zhang et al., 2003)—thus raising the question: can one assume that the mode of chloroplast inheritance is identical among congeners? If not, then chloroplast inheritance needs to be assessed directly in each species in question.
Several factors influence whether paternal leakage can occur and what effect it has on the population biology of plant species: whether and how often pollen grains contain cpDNA, how frequently it is transmitted to the zygote, and how intraindividual drift affects copies within cells. Large-scale studies designed to infer organellar inheritance in angiosperms have screened pollen grains for evidence of plastids or plastid DNA in generative or sperm cells (Sears, 1980; Corriveau and Coleman, 1988; Zhang et al., 2003). Using cytological evidence to determine the mode of plastid DNA inheritance, these studies have designated cpDNA transmission in hundreds of angiosperm species as either maternal or biparental. Due to their nature, these methods can only identify the potential mode of inheritance, that is, whether there is plastid DNA in the cell. Since these studies scan a great number of species, they often use a small number of individuals per species, which may miss rare paternal transmission. It is also not possible to determine the consequences for individual and population biology when the transmission mode is biparental since intraindividual drift of DNA copies during cell divisions will lead to individuals that sort almost completely to one type or the other (that is, a homoplasmic individual) or to individuals that carry a mixture of paternal and maternal copies (that is, a heteroplasmic individual) (Birky, 2001). Cell divisions represent founder effects or bottlenecks of each generation, which should enforce a highly homoplasmic state within the individual. However, occasional biparental inheritance would continue to introduce alternate alleles into the individual generating heteroplasmy. Chloroplast heteroplasmy has been documented in several angiosperms genera: Passiflora (Hansen et al., 2006, 2007), Senecio (Frey et al., 2005), Medicago (Johnson and Palmer, 1989) and Turnera (Shore et al., 1994; Shore and Triassi, 1998).
We have investigated cpDNA inheritance and heteroplasmy in a rare sunflower species, Helianthus verticillatus Small, and compared it to the mode of inheritance found in a related economically important species, H. annuus (Rieseberg et al., 1994; Wills et al., 2005). These two prior studies found no evidence for paternal leakage of cpDNA in H. annuus crosses, and Wills et al. (2005) found no evidence for heteroplasmy (personal communication). Ellis et al. (2006) investigated population structure in H. verticillatus based on FST calculations for nuclear and cpDNA markers and found the latter to have a much greater FST. On the basis of the described cpDNA inheritance in H. annuus, the difference in magnitude between nuclear and chloroplast FST was explained in part by maternal inheritance of cpDNA. However, this assumed mode of inheritance has not been tested directly in H. verticillatus. Since we are focusing on just one species, we chose to look at chloroplast inheritance directly by examining progeny from controlled crosses in which the parents had different cpDNA haplotypes. This approach allows the direct observation of chloroplast transmission and includes much larger sample sizes, thus giving a greater chance of detecting rare paternal leakage and quantifying the rate of leakage accurately.
When designing an experiment to study rare events, one must take into account the power of detection. Milligan (1992) proposed a binomial model of organelle inheritance to determine the power of the analysis to detect paternal leakage at a given rate. He states that many studies that address organelle inheritance use insufficient sample sizes to detect leakage, and since some studies have found leakage rates of 0.01–2.5% (Simmonds, 1969; Tilney-Bassett, 1978; Medgyesy et al., 1986), sample sizes in excess of 100 progeny are needed to detect leakage even at the 2.5% level. Further, since the probability of non-maternal chloroplast inheritance may vary among crosses (Birky, 1995; Mogensen, 1996), individuals will not be completely independent data points if they are from the same family. Consequently, when designing an experiment to evaluate organelle inheritance, one should choose as many families as possible recognizing the trade-off between the number of families and the number of offspring per family. Here we report on a study of cpDNA inheritance using cpSSRs (chloroplast simple sequence repeat; Provan et al., 2001) in 323 H. verticillatus offspring comprising 53 families and provide evidence for occasional paternal leakage and heteroplasmy of cpDNA in controlled greenhouse crosses.
Materials and methods
H. verticillatus is an extremely rare, diploid (n=17) self-incompatible, perennial sunflower restricted to only four locations in the southeast interior of the United States: two in western Tennessee (Madison Co. and McNairy Co.), one in northeastern Alabama (Cherokee Co.) and one in northwestern Georgia (Floyd Co.). It is a candidate for federal listing for the Endangered Species Act and is listed as endangered in each of the three states. First collected in western Tennessee in 1892, it was not found again in the field until 1994 in Georgia (Matthews et al., 2002). In 1996 and 1998, the populations in Alabama and Madison Co., Tennessee, respectively, were discovered. In the fall of 2006, the fourth location in McNairy Co., Tennessee was discovered during an annual survey and search for the species. The Alabama and Georgia populations are about 3.5 km from each other, whereas the Tennessee populations are about 350 km from the others and about 40 km from one another. In a prior study, the Alabama and Tennessee populations were found to be fixed for different cpDNA haplotypes making it possible to detect paternal leakage easily in crosses between the two populations (Ellis et al., 2006).
In order to detect rare paternal leakage, the appropriate sample size must be used. We used Milligan's (1992) equation for calculating the power of analysis for the number of individuals studied and the allowed percentage of non-maternal inheritance:

where β is the power of the test to detect leakage, P is the probability of paternal transmission, and N is the number of progeny. We designed our experiment to be able to reject the strict maternal inheritance hypothesis 95% of the time at a rate of leakage equal to or >1%. To have this statistical power, 300 individuals (that is, observations of inheritance) were needed according to this calculation.
Achenes from H. verticillatus were collected from the Alabama and Tennessee sites and grown in the Vanderbilt Biological Sciences greenhouse to serve as parents for the crosses. Since the parents need to differ at the markers studied in order to detect paternal leakage, we genotyped the parents for the cpSSRs (method described below) to choose individuals with differing cpDNA haplotypes. Alabama and Tennessee individuals carried different cpDNA haplotypes; therefore, any interpopulation crosses would have differing parental genotypes. The Tennessee population was polymorphic; thus, Tennessee by Tennessee intrapopulation crosses were also conducted with parents that differed in cpDNA haplotype. We used 272 offspring from 45 controlled greenhouse interpopulation crosses (24 Alabama × Tennessee and 21 Tennessee × Alabama) and 51 individuals from eight greenhouse intrapopulation (Tennessee) crosses of H. verticillatus for a total of 323 offspring.
The crosses were carried out as follows: inflorescences were bagged prior to anthesis to prevent any unwanted pollinations. Crosses were conducted by brushing pollen with a paintbrush from inflorescences at anthesis into aluminum foil and then brushing pollen onto the stigmas of another inflorescence in which the same pollen removal had been conducted. Pollinations were conducted within 1 h of collecting pollen and all pollinations were conducted at mid-morning (∼1000 h). Inflorescences were re-bagged and achenes were allowed to mature. Achenes (that is, the offspring) were nicked with a razor blade, germinated on moist filter paper and grown in the greenhouse. When the resulting plants were large enough, a leaf was collected for DNA extraction.
Total genomic DNA was isolated from ∼200 mg of fresh leaf tissue using the Applied Biosystems 6100 Nucleic Acid PrepStation DNA extraction machine and associated protocols (Foster City, CA, USA). All parents and offspring were genotyped for three polymorphic cpSSRs, (N39 and N30 (Bryan et al., 1999) and C7 (Weising and Gardner, 1999); also used in Wills et al., 2005; Ellis et al., 2006) using PCR and fragment length analysis. Any individuals that indicated paternal leakage were re-genotyped two additional times to verify the results. Briefly, SSR genotyping was performed using a modified version of the fluorescent labeling protocol of Schuelke (2000), as detailed in Wills et al. (2005). PCR was performed in a total volume of 20 μl containing 2 ng of template DNA, 30 mM Tricine pH 8.4-KOH, 50 mM KCl, 2 mM MgCl2, 125 μM of each deoxyribonucleotide triphosphate, 0.2 μM M13 Forward (−29) sequencing primer labeled with either HEX, 6FAM or TET, 0.2 μM reverse primer, 0.02 μM forward primer and two units of Taq polymerase. The PCR conditions were as follows: 3 min at 95 °C; ten cycles of 30 s at 94 °C, 30 s at 65 °C and 45 s at 72 °C, annealing temperature decreasing to 55 °C by 1 °C per cycle, followed by 30 cycles of 30 s at 94 °C, 30 s at 55 °C, 45 s at 72 °C, followed by 20 m at 72 °C.
PCR products were visualized on an MJ Research BaseStation automated DNA sequencer (South San Francisco, CA, USA). MapMarker 1000 ROX size standards (BioVentures Inc., Murfreesboro, TN, USA) were run in each lane to allow for accurate determination of fragment size. Cartographer v 1.2.6 (MJ Research) was used to infer individual genotypes according to the fragment sizes of the PCR products. The parents used in this study carried one of four haplotypes with Alabama and Tennessee containing different haplotypes. Haplotypes were denoted A1, T1, T2, T3 named by the three-locus SSR sizes (C7–N30–N39: 145–176–174 bp=A1; 148–177–181 bp=T1; 149–177–184 bp=T2; 149–177–185 bp=T3). Given that individuals showing paternal leakage would likely still carry some of the maternal haplotype (that is, heteroplasmy), we scored individuals first for their primary fragment peak and scored any secondary peak when it met two criteria: (1) it was the alternate allele that would be expected given the type of cross and (2) it was at least 10% of the primary peak fluorescence level. Any individual that indicated heteroplasmy was also genotyped two additional times (from the same DNA extraction). All parents were found to be of one primary haplotype.
Since paternal leakage is most likely a rare occurrence, it is critical that parentage be verified to ensure that mistakes were not made during the handling of individuals throughout cultivation, DNA extractions and genotyping. In order to detect any possible errors in mother–offspring assignment, parentage was verified in those offspring that did not indicate maternal cpDNA inheritance by genotyping the suspected leakage individuals as well as the maternal and paternal donors for nine previously described highly polymorphic nuclear EST-SSRs BL 1, 2, 3, 4, 5, 8, 10, 13 and 17 (for details see Pashley et al., 2006; Ellis et al., 2006).
Results
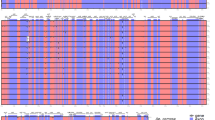
Out of 323 observations of inheritance, we found five cases of non-maternal inheritance equaling a leakage rate of at least 1.55%. Table 1 provides the cpDNA haplotypes for the offspring indicating paternal leakage. One offspring each of the AL2 × TN2, TN4 × AL3, TN6 × AL3 families showed the paternal haplotype. Two individuals of the TN3 × AL2 family had the paternal haplotype. Maternity and paternity was confirmed in each case that indicated paternal leakage using EST-SSRs. In each of the 323 offspring scored, a primary haplotype was observed; however, some offspring contained a secondary haplotype (at least 10% of the primary peak) that represented the alternate allele of the appropriate size for the cross type (see Figure 1 for a representative example). The highest ratio observed for primary to secondary haplotype was roughly 2:1. All five individuals that indicated primary non-maternal inheritance also carried at least 10% of the maternal haplotype, and one additional individual (from family AL2 × TN2) primarily contained the maternal haplotype but contained an observable percentage of the paternal haplotype according to our criteria (Table 2). Further, a Fisher's Exact Test indicated that the probability of detecting heteroplasmy is not independent of the probability of which parent provides the primary haplotype (P<0.001).
Electropherogram from cpSSR (chloroplast simple sequence repeat) locus N39 to show an example of chloroplast inheritance. A mother and three offspring from the AL2 × TN2 family are shown. Offspring one and two show maternal inheritance, while offspring three contains the paternal allele as well as evidence of the maternal allele, indicating paternal leakage and biparental inheritance. There is a size marker at the 200 bp position.
Discussion
In this study we found paternal leakage of the chloroplast at a rate of 1.55% (5/323 observations) or 1.86% (6/323) accounting for the individual that showed secondary paternal heteroplasmy. This low level of paternal transmission was observable in our study since we designed the experiment to be able to detect leakage 95% of the time at a transmission rate of ⩾1% (Milligan, 1992). One caveat to discuss with regard to calculating the detection ability is that since there is sometimes a family association with leakage (Birky, 1995; Mogensen, 1996; McCauley et al., 2007), the actual number of independent data points may not be the total number of observations, but rather probably lies somewhere between the number of families and the number of individuals. Thus when approaching the trade-off between number of families and offspring, and given a constraint on the total number of samples in a study, it is better to sample as many families as possible. Studies that employ a small number of families to detect non-maternal inheritance may be more likely to miss rare events of paternal leakage. In this study we sampled 53 families—a number that is much larger than previous studies finding strict maternal chloroplast inheritance in other angiosperms (Vaillancourt et al., 2004; Van Droogenbroeck et al., 2005). We only found evidence for paternal leakage in interpopulation crosses. Often paternal leakage is reported in interspecific hybrids (Soliman et al., 1987; Cruzan et al., 1993; Hansen et al., 2007) and thus it may be more likely for paternal leakage to occur in crosses between divergent populations than within population crosses. However, no formal statistical calculations were performed on this conclusion as we had so few within population crosses, thus limiting our statistical power.
We also found evidence for chloroplast heteroplasmy in six individuals. Documentation of chloroplast heteroplasmy is rare (but see Frey et al., 2005) perhaps due in part to the dogma of strict maternal inheritance in angiosperms. Intraindividual variation has been observed and quantified for plant mitochondrial genes in several cases, (Hattori et al., 2002; McCauley et al., 2005; Welch et al., 2006) but such observations remain rare. Perhaps not surprisingly heteroplasmy occurs if paternal leakage takes place given that the mother presumably always transfers organelles to her offspring. Chloroplast heteroplasmy has consequences for the population biology of H. verticillatus. Given that founder effects or bottlenecks of chloroplasts occur with each successive cell division, even a small amount of paternal leakage at fertilization could, by chance, lead to a mature offspring with the majority paternal haplotype just as it might often be lost. Genetic drift within the individual will also vary at different life stages of the plant. A young plant will likely have fewer cell divisions and may harbor a greater mixture of chloroplast genes, while an older plant has completed more cell divisions thus allowing genetic drift to create a more homoplasmic state (recall we sampled H. verticillatus offspring at the young plant stage).
The findings of this study indicate caution must be used when assuming strict maternal inheritance of the chloroplast genome in angiosperms and in unstudied species of Helianthus for several reasons. First, cpDNA is often employed in studies examining hybridization and introgression in plants (Rieseberg and Ellstrand, 1993; Edwards-Burke et al., 1997; Welch and Rieseberg, 2002; Van Droogenbroeck et al., 2006). When strict maternal inheritance is assumed but paternal leakage occurs, incorrect conclusions regarding parental contributions during hybridization and directionality of introgression may be drawn. The sunflower genus is noted for having significant amounts of hybridization and introgression (Rieseberg, 1991; Rieseberg et al., 1995, 1996). Paternal transmission of the chloroplast found in H. verticillatus indicates the need to be cautious when studying aspects of hybridization in this genus.
Second, non-maternal inheritance may complicate cpDNA-based phylogenies since chloroplast transmission through pollen would increase the likelihood of its introgression from one species into another. This ‘chloroplast capture’ can result in incongruence between nuclear and organelle phylogenies (Soltis and Kuzoff, 1995; Tsitrone et al., 2003). Schilling et al. (1998) found inconsistencies between nuclear and cpDNA phylogenies in Helianthus. Since paternal transmission of the chloroplast could lead to inconsistencies when comparing nuclear and chloroplast phylogenies, our findings suggest that the possibility of paternal leakage should be considered when examining evolutionary relationships in Helianthus.
Next, the difference between measures of population structure (FST) using cpDNA and nuclear DNA is often used to evaluate the relative contributions of seed and pollen movement to total gene flow. In theory, if there is strict maternal inheritance of organellar genes then seeds will carry copies of the nuclear and cytoplasmic genomes, while pollen will carry only nuclear genes (Birky et al., 1983, 1989; Petit et al., 1993). Paternal leakage will decrease values of FST based on organellar DNA, skewing estimates of the contributions of seeds and pollen to gene flow relative to that when maternal inheritance is assumed.
Finally, it has been proposed that transgenes be placed in the chloroplast genome of crop species to prevent their escape (Gressel, 1999; Daniell et al., 2005; Grevich and Daniell, 2005). However, low rates of paternal transmission of the chloroplast have been shown in crops species including tobacco (Medgyesy et al., 1986) and potato (Simmonds, 1969). Haygood et al. (2004) modeled transgene escape and found that even genes with leakage rates as low as that found in our study will have an appreciable probability of escape into the wild. The possibility of transgene escape at such low leakage rates further highlights the necessity for studies examining mode of organellar inheritance to have a high statistical power to detect rare leakage.
We were also interested in addressing the question of whether it is necessary to study chloroplast transmission in more than one species within the same genus (that is, is it possible to extrapolate within the genus if one member species' transmission has already been determined?). In order to address this, we considered the results of three surveys of potential chloroplast transmission in angiosperms (Sears, 1980; Corriveau and Coleman, 1988; Zhang et al., 2003) and after accounting for overlap among them and assuming equal sampling intensities, we found that 6 out of 113 genera contained conflicting modes of inheritance. In three of the genera, there was predominantly one type of transmission with a low frequency of the other. In our study, we found H. verticillatus to have a low level of paternal leakage. This is in contrast to the observation of strict maternal inheritance found in another species, H. annuus, within the genus (Rieseberg et al., 1994; Wills et al., 2005). Combining their data set with that of Rieseberg et al. (1994), Wills et al. (2005) determined they were able to detect paternal leakage at a rate of ⩾1.35%, 95% of the time, a study that is comparable in magnitude to ours. The findings of this literature survey and our results indicate that the mode of chloroplast inheritance cannot always be extrapolated within a genus. Furthermore, the methods used in these studies may only provide conservative estimates given the limited sample sizes per species and ability to detect only the potential mode of inheritance.
In conclusion, we found evidence for a modest amount paternal transmission of cpDNA and heteroplasmy in the perennial sunflower, H. verticillatus, in greenhouse crosses. We are also interested in the consequences of paternal leakage and heteroplasmy for the biology of natural populations. The next step is to address paternal leakage in the field. If we extrapolate our primary paternal leakage rate (1.55%) to natural populations and assume every leakage event is detectable (that is, the parents have different cpDNA haplotypes), at least 200 offspring from the field would be necessary to have a 95% chance of detecting leakage at the frequency we found in the crosses. However, every leakage event will not be detectable given the high level of chloroplast population structure, FST=0.620 (Ellis et al., 2006). The probability of the parents being different, assuming random mating within populations, can be calculated for n number of haplotypes by

where p, q, r and s are cpDNA haplotype frequencies from a given natural population. For example, using the four haplotype frequencies from the Tennessee population of H. verticillatus (Ellis et al., 2006), P=0.48 (1−0.72−0.122−0.122−0.062). Roughly, only half of the matings will be between parents that differ at these cpSSRs. Thus, multiplying 200, the number of offspring that must be inspected to observe leakage at a rate of 1.55 when parents differ at marker loci, by 1/P yields the expectation that 417 offspring must be examined to have a 95% chance of detecting leakage at a rate of 1.55% in natural populations given the population structure. These results indicate large sample sizes are necessary to ensure detection of rare paternal leakage in angiosperms especially in natural populations.
References
Birky CW (1995). Uniparental inheritance of mitochondrial and chloroplast genes: mechanisms and evolution. Proc Nat Acad Sci USA 92: 11331–11338.
Birky CW (2001). The inheritance of genes in mitochondria and chloroplasts: laws, mechanisms, and models. Annu Rev Gen 35: 125–148.
Birky CW, Fuerst P, Maruyama T (1989). Organelle gene diversity under migration, mutation, and drift—equilibrium expectations, approach to equilibrium, effects of heteroplasmic cells, and comparison to nuclear genes. Genetics 121: 613–627.
Birky CW, Maruyama T, Fuerst P (1983). An approach to population and evolutionary genetic theory for genes in mitochondria and chloroplasts, and some results. Genetics 103: 513–527.
Bryan GJ, McNicoll J, Ramsay G, Meyer RC (1999). Polymorphic simple sequence repeat markers in chloroplast genomes of Solanaceous plants. Theor Appl Genet 99: 859–867.
Chapman MA, Burke JM (2006). Letting the gene out of the bottle: the population genetics of genetically modified crops. New Phytol 170: 429–443.
Corriveau JL, Coleman AW (1988). Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. Am J Bot 75: 1443–1458.
Cruzan MB, Arnold ML, Carney SE, Wollenberg KR (1993). cpDNA inheritance in interspecific crosses and evolutionary inference in Louisiana Irises. Am J Bot 80: 344–350.
Daniell H, Kumar S, Dufourmantel N (2005). Breakthrough in chloroplast genetic engineering of agronomically important crops. Trends Biotechnol 23: 238–245.
Dobes̆ CH, Mitchell-Olds T, Koch MA (2004). Extensive chloroplast haplotype variation indicates Pleistocene hybridization and radiation of North American Arabis drummondii, A. x divaricarpa, and A-holboellii (Brassicaceae). Mol Ecol 13: 349–370.
Edwards-Burke MA, Hamrick JL, Price RA (1997). Frequency and direction of hybridization in sympatric populations of Pinus taeda and P. echinata (Pinaceae). Am J Bot 75: 1143–1458.
Ellis JR, Pashley CH, Burke JM, McCauley DE (2006). High genetic diversity in a rare and endangered sunflower as compared to a common congener. Mol Ecol 15: 2345–2355.
Frey JE, Frey B, Forcioli D (2005). Quantitative assessment of heteroplasmy levels in Senecio vulgaris chloroplast DNA. Genetica 123: 255–261.
Gressel J (1999). Tandem constructs: preventing the rise of superweeds. Trends Biotechnol 17: 361–366.
Grevich JJ, Daniell H (2005). Chloroplast genetic engineering: recent advances and future perspectives. Crit Rev Plant Sci 24: 83–107.
Hamilton MB, Miller JR (2002). Comparing relative rates of pollen and seed gene flow in the island model using nuclear and organelle measures of population structure. Genetics 162: 1897–1909.
Hansen AK, Escobar LK, Gilbert LE, Jansen RK (2007). Paternal, maternal, and biparental inheritance of the chloroplast genome in Passiflora (Passifloraceae): implications for phylogenetic studies. Am J Bot 94: 42–46.
Hansen AK, Gilbert LE, Simpson BB, Downie SR, Cervi AC, Jansen RK (2006). Phylogenetic relationships and chromosome number evolution in Passiflora. Syst Bot 31: 138–150.
Hattori N, Kitagawa K, Takumi S, Nakamura C (2002). Mitochondrial DNA heteroplasmy in wheat, Aegilops and their nucleus-cytoplasm hybrids. Genetics 160: 1619–1630.
Haygood RA, Ives AR, Andow DA (2004). Population genetics of transgene containment. Ecol Lett 7: 213–220.
Johnson LB, Palmer JD (1989). Heteroplasmy of chloroplast DNA in Medicago. Plant Mol Biol 12: 3–11.
Kelchner SA (2000). The evolution of non-coding chloroplast DNA and its application to plant systematics. Ann Mo Bot Gard 87: 482–498.
Matthews JF, Allison JR, Ware RT, Nordman C (2002). Helianthus verticillatus Small (Asteraceae) rediscovered and redescribed. Castanea 67: 13–24.
McCauley DE (1995). The use of chloroplast DNA polymorphism in studies of gene flow in plants. Trends Ecol Evol 10: 198–202.
McCauley DE, Bailey MF, Sherman NA, Darnell MZ (2005). Evidence for paternal transmission and heteroplasmy in the mitochondrial genome of Silene vulgaris, a gynodioecious plant. Heredity 95: 50–58.
McCauley DE, Sundby AK, Bailey MF, Welch ME (2007). The inheritance of chloroplast DNA is not strictly maternal in Silene vulgaris (Caryophyllaceae): evidence from experimental crosses and natural populations. Am J Bot 94: 1333–1337.
Medgyesy P, Pay A, Marton L (1986). Transmission of paternal chloroplasts in Nicottana. Mol Gen Genet 204: 195–198.
Milligan BG (1992). Is organelle DNA strictly maternally inherited? Power analysis of a binomial distribution. Am J Bot 79: 1325–1328.
Mogensen HL (1996). The hows and whys of cytoplasmic inheritance in seed plants. Am J Bot 83: 383–404.
Olmstead RG, Palmer JD (1994). Chloroplast DNA systematics: a review of methods and data analysis. Am J Bot 81: 1205–1224.
Ouborg NJ, Piquot Y, Van Groenendael JM (1999). Population genetics, molecular markers and the study of dispersal in plants. J Ecol 87: 551–568.
Pashley CH, Ellis JR, McCauley DE, Burke JM (2006). EST databases as a source for molecular markers: lessons from Helianthus. J Heredity 97: 381–388.
Petit RJ, Duminil J, Fineschi S, Hampe A, Salvinin D, Vendramin GG (2005). Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol Ecol 14: 689–701.
Petit RJ, Kremer A, Wagner DB (1993). Finite island model for organelle and nuclear genes in plants. Heredity 71: 630–641.
Provan JW, Powell W, Hollingsworth PM (2001). Chloroplast microsatellites: new tools for studies in plant ecology and evolution. Trends Ecol Evol 16: 142–147.
Reboud X, Zeyl C (1994). Organelle inheritance in plants. Heredity 72: 132–140.
Rieseberg LH (1991). Homoploid reticulate evolution in Helianthus (Asteraceae): evidence from ribosomal genes. Am J Bot 78: 1218–1237.
Rieseberg LH, Ellstrand NC (1993). What can molecular and morphological markers tell us about plant hybridization? Crit Rev Plant Sci 12: 213–241.
Rieseberg LH, Sinervo B, Linder CR, Ungerer MC, Arias DM (1996). Role of gene interactions in hybrid speciation: evidence from ancient and experimental hybrids. Science 272: 741–745.
Rieseberg LH, Van Fossen C, Arias D, Carter RL (1994). Cytoplasmic male-sterility in sunflower-origin, inheritance, and frequency in natural populations. J Heredity 85: 233–238.
Rieseberg LH, Van Fossen C, Desrochers AM (1995). Hybrid speciation accompanied by genomic reorganization in wild sunflowers. Nature 375: 313–316.
Röhr H, Kues U, Stahl U (1998). Organelle DNA of plants and fungi: inheritance and recombination. Prog Bot 60: 39–87.
Schilling EE, Linder CR, Noyes RD, Rieseberg LH (1998). Phylogenetic relationships in Helianthus (Asteraceae) based on nuclear ribosomal DNA internal transcribed spacer region sequence data. Syst Bot 23: 177–187.
Schuelke M (2000). An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol 18: 233–234.
Sears BB (1980). Elimination of plastids during spermatogenesis and fertilization in the plant kingdom. Plasmid 4: 233–255.
Shore JS, McQueen KL, Little SH (1994). Inheritance of plastid DNA in the Turnera-ulmifolia complex (Turneraceae). Am J Bot 81: 1636–1639.
Shore JS, Triassi M (1998). Paternally biased cpDNA inheritance in Turnera ulmifolia (Turneraceae). Am J Bot 85: 328–332.
Simmonds NW (1969). Variegated mutant plastid chimeras of potatoes. Heredity 24: 303–306.
Smith SE (1989). Biparental inheritance of organelles and its implications for crop improvement. Plant Breed Rev 6: 361–393.
Soliman K, Fedak G, Allard RW (1987). Inheritance of organelle DNA in Barley and Hordeum × Secale intergeneric hybrids. Genome 29: 867–872.
Soltis DE, Kuzoff RK (1995). Discordance between nuclear and chloroplast phylogenies in the Heuchera group (Saxifragaceae). Evolution 49: 727–742.
Tilney-Bassett RAE (1978). The inheritance and genetic behavior of plastids. In: Kirk JTO, Tilney-Bassett RAE (eds). The Plastids. Their Chemistry, Structure, Growth and Inheritance. Elsevier: New York, NY. pp 251–524.
Tsitrone A, Kirkpatrick M, Levin DA (2003). A model for chloroplast capture. Evolution 57: 1776–1782.
Vaillancourt RE, Petty A, McKinnon GE (2004). Maternal inheritance of mitochondria in Eucalyptus globulus. J Heredity 95: 353–355.
Van Droogenbroeck B, Kyndt T, Romeijn-Peeters E, Van Thuyne W, Goetghebeur P, Romero-Motochi JP et al. (2006). Evidence of natural hybridization and introgression between Vasconcellea species (Caricaceae) from southern Ecuador revealed by chloroplast, mitochondrial and nuclear DNA markers. Ann Bot 97: 793–805.
Van Droogenbroeck B, Maertens I, Haegeman A, Kyndt T, O'brien C, Drew RA et al. (2005). Maternal inheritance of cytoplasmic organelles in intergeneric hybrids of Carica papaya L. and Vasconcellea spp. (Caricaceae Dumort., Brassicales). Euphytica 143: 161–168.
Weising K, Gardner RC (1999). A set of conserved PCR primers for the analysis of simple sequence repeat polymorphisms in chloroplast genomes of dicotyledonous angiosperms. Genome 42: 9–19.
Welch ME, Darnell MZ, McCauley DE (2006). Variable populations within variable populations: quantifying mitochondrial heteroplasmy in natural populations of the gynodioecious plant Silene vulgaris. Genetics 174: 829–837.
Welch ME, Rieseberg LH (2002). Habitat divergence between a homoploid hybrid sunflower species, Helianthus paradoxus (Asteraceae), and its progenitors. Am J Bot 89: 472–478.
Wills DM, Hester ML, Liu AZ, Burke JM (2005). Chloroplast SSR polymorphisms in the Compositae and the mode of organellar inheritance in Helianthus annuus. Theor Appl Genet 110: 941–947.
Wolfe AD, Randle CP (2004). Recombination, heteroplasmy, haplotype polymorphism, and paralogy in plastid genes: Implications for plant molecular systematics. Syst Bot 29: 1011–1020.
Zhang Q, Liu Y, Sodmergen (2003). Examination of the cytoplasmic DNA in male reproductive cells to determine the potential for cytoplasmic inheritance in 295 angiosperm species. Plant Cell Phys 44: 941–951.
Acknowledgements
We thank Charles and Mary Lipscomb and Jonathan Ertelt for their assistance in the greenhouse.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ellis, J., Bentley, K. & McCauley, D. Detection of rare paternal chloroplast inheritance in controlled crosses of the endangered sunflower Helianthus verticillatus. Heredity 100, 574–580 (2008). https://doi.org/10.1038/hdy.2008.11
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2008.11
Keywords
This article is cited by
-
Paternal leakage inheritance and a fitness cost are associated with the chloroplastic psbA gene controlled metribuzin tolerance in lentil (Lens culinaris)
Euphytica (2021)
-
Rare maternal and biparental transmission of the cucumber mitochondrial DNA reveals sorting of polymorphisms among progenies
Theoretical and Applied Genetics (2019)
-
Natural history of the narrow endemics Ipomoea cavalcantei and I. marabaensis from Amazon Canga savannahs
Scientific Reports (2017)
-
Maternal inheritance of mitochondrial genomes and complex inheritance of chloroplast genomes in Actinidia Lind.: evidences from interspecific crosses
Molecular Genetics and Genomics (2013)
-
Comparative analysis of the complete sequence of the plastid genome of Parthenium argentatum and identification of DNA barcodes to differentiate Parthenium species and lines
BMC Plant Biology (2009)