Abstract
Purpose:
Rare genetic variants are the major cause of Mendelian disorders, yet only half of described genetic diseases have been causally linked to a gene. In addition, the total number of rare genetic diseases is projected to be far greater than that of those already described. Whole-genome sequencing of patients with subsequent genetic and functional analysis is a powerful way to describe these gene anomalies. However, this approach results in tens to hundreds of candidate disease-causative genes, and the identification of additional individuals suffering from the same disorder can be difficult because of rarity and phenotypic heterogeneity.
Methods:
We describe a genetic network–based method to rank candidate genes identified in family-based sequencing studies, termed phenotype informed network (PIN) ranking. Furthermore, we present a case study as an extension of the PIN ranking method in which disease symptoms drive the network ranking and identification of the disease-causative gene.
Results:
We demonstrate, through simulation, that our method is capable of identifying the correct disease-causative gene in a majority of cases. PIN-rank is available at https://genomics.scripps.edu/pinrank/.
Conclusion:
We have developed a method to prioritize candidate disease-causative genes based on symptoms that would be useful for both the prioritization of candidates and the identification of additional subjects.
Genet Med 17 11, 859–865.
Similar content being viewed by others
Introduction
Rare and de novo genetic variants are a major cause of Mendelian disorders. As the average age of first-time parents continues to increase, the burden of rare variants in the population and the incidence of rare Mendelian diseases caused by de novo variants is also expected to increase.1 Of the rare Mendelian diseases that have already been described, approximately half have yet to be linked to a causal gene (idiopathic diseases).2 Estimates of the total number of rare Mendelian diseases based on the number of known disease-causing and essential genes have resulted in predictions of between 7,000 and 15,000 total rare Mendelian diseases, suggesting many have yet to be described.3
High-throughput genome sequencing has provided a powerful tool for the identification of genetic mutations underlying rare Mendelian diseases.4,5,6,7 The general approach to providing a genetic diagnosis for idiopathic diseases involves a combination of classic genetic analysis, various variant filters, and literature-backed assessments of the likely functional consequences of individual genetic variants.8,9 This combination of techniques, typically applied to a nuclear family, or trio, can result in tens to hundreds of candidate variants that may underlie a rare disease, depending on the segregation pattern of the disease through the trio and the stringency of applied filters. For diseases that do not fit a previously described indication of a known disease because of the well-known phenotypic heterogeneity underlying rare Mendelian diseases,10 confident prioritization and discovery of novel disease-causative variants can be problematic. Although any gene–disease association should ultimately be confirmed by segregation in multiple families and/or occurrence in multiple unrelated individuals with the same phenotype,11 for extremely rare and/or heterogeneous phenotypes, prioritizing candidate variants in single families based on symptoms or related diseases is useful to guide functional validation efforts. Although some tools exist for prioritizing variants in known disease-causing genes based on phenotype or other information,12,13 few exist for the identification of novel gene–disease relationships (see Discussion). Toward this end, we propose an approach that merges classic genetic analysis with a genetic network–based prioritization approach for the identification of novel disease-causative genes. In addition, we describe an extension of this approach driven directly by the symptoms exhibited by an individual with disease. We term this approach “phenotype-informed network ranking,” or “PIN rank.”
Materials and Methods
Test genomes
Twenty-two trios were collected to simulate disease gene identification in a family-based genome sequencing setting. Whole exomes for 12 of the trios were obtained from the 1000 Genomes Project,14 and the remainder were obtained through the Scripps Translational Science Institute Idiopathic Diseases of Man protocol (IDIOM trios; Supplementary Table S1 online). Written informed consent was acquired from all adult members of the IDIOM trios according to the protocol approved by the Scripps Health institutional review board.
Sequencing and variant calling for the 1000 Genomes trios was performed by Complete Genomics (ftp://ftp.1000genomes.ebi.ac.uk/) using version 2.2.0.19 of its variant-calling pipeline.15 For the IDIOM trios, genomic DNA was collected from whole-blood samples using the QIAamp system (Qiagen, Valencia, CA). Enriched exome libraries were prepared using the Agilent Sureselect XT kit (Agilent, Santa Clara, CA). Whole-exome sequences were obtained at Scripps Translational Science Institute on the Illumina HiSeq2000 platform with a 100-bp, indexed, paired-end sequencing run (TruSeq SBS Kit v3 200 cycle (Illumina, San Diego, CA)). A mean coverage of between 79× and 119× was achieved per individual, with 93–95% of the target exome covered by more than 10 reads in all individuals. Sequence alignment and variant calling were performed against the reference human genome (National Center for Biotechnology Information 37/hg 19), and variant calling was done using a best-practices BWA-GATK Unified Genotyper pipeline.16,17,18
Test genome filtering
Variants in each trio were filtered under the assumption that the disease segregated in each family as a simple Mendelian trait, that is, it results from a de novo dominant mutation or is inherited in a simple recessive or compound heterozygous manner. Specifically, all the variants were first run through the SG-ADVISER pipeline (https://genomics.scripps.edu/ADVISER/). We then filtered the results to retain all the variants affecting protein coding (nonsynonymous, in-frame, frameshift, and nonsense variants) and/or splicing in the University of California–Santa Cruz Known Genes table and that had less than 1% allele frequency in each reference population (HapMap,19 1000 Genomes,14 National Heart, Lung, and Blood Institute,20 or Wellderly21,22 cohorts), and which segregated in the family under the assumption of unaffected parents and an affected proband—the most common scenario encountered in our IDIOM cases. For de novo variants, we further filtered variants to retain only those present at less than 0.1% allele frequency to remove false de novo calls caused by variant-calling errors. The filtering resulted in a set of genes affected by rare genetic variants, and this set was used as candidate disease-causative genes for our simulations described below.
Implanted disease-causative variants
The Human Gene Mutation Database (version 2013.3) was downloaded and used to build our test set of disease-causative genes (implanted genes) and to select the seeds, that is, the genes associated with the symptoms of a diseased individual, for the network-based ranking algorithm. Human Gene Mutation Database variants were run through the SG-ADVISER pipeline and filtered to remove potential false-positive entries; variants with a frequency <1% in the HapMap, 1000 Genomes, and National Heart, Lung, and Blood Institute exomes and Wellderly data were retained. Of the remaining variants, a test set was constructed from disease-causative variants mapped to genes that had not been associated with any disease before 2011. The set of seed genes consisted of all genes that had been associated with the diseases represented in the test set before 2011. As described below, our genetic network is derived from data available before 2011—this temporal separation is crucial to accurately replicating the discovery of novel gene–disease relationships.
Phenotype informed ranking
PIN-rank operates by ranking candidate disease–causing genes based on the fold change in their basic PageRank23 versus the phenotype-informed personalized PageRank24,25 within a genetic network. Both of these ranks are obtained by iterating the function (Ri+1 = αARi + (1 − α)T) for different values of α and T until convergence.
Assuming the total number of genes in the network is n, A is a weighted directed adjacency matrix of dimension (n,n) containing the information regarding how genes are linked in the genetic network, T is a teleportation matrix of dimension (n,1) containing the probabilities of randomly teleporting to each gene in the genetic network, α is an adjustable factor denoting how often one moves along the links within the adjacency matrix versus teleporting to genes within the genetic network, and Ri is a matrix of the PageRanks of dimension (n,1), or the equilibrium probability that one will arrive at each gene by following the links within the adjacency matrix or teleporting. The final value of R (Ri) is arrived at by initializing R with equal probabilities for all genes and solving by the power method, or iterating the above calculation until R stabilizes (Ri+1 − Ri < 1 × 10−8).
Genetic network
The values within the adjacency matrix A (n = 17,369 genes for our simulations) were derived from the probability with which each gene is connected to another in the StringDB version 8.3 database.26 StringDB integrates genomic context, known protein–protein interactions, coexpression, and literature mining to derive these probabilities. Version 8.3 was chosen so that information gathered after 2011—that is, after the discovery of our test disease genes—was not contained within the network. For PIN-rank, we created three separate adjacency matrices: one with unscaled StringDB probabilities and two with StringDB probabilities scaled to the second or third power. The latter two adjacency matrices down-weight the low-probability links. Path lengths within this network were calculated using the iGraph library (version 0.6.5) (ref. 27) implementation of Dijkstra’s algorithm, in which connection lengths were defined as the inverse of the weight of the connection between the genes.
Basic and personalized PageRank calculations
For basic PageRank, α is set at 0.99 and T is set at equal probabilities for every gene, effectively removing any effect of teleportation on the ranks of genes within the network while allowing R to stabilize in the face of dangling nodes or other factors known to disrupt R stabilization via the power method. For the phenotype-informed personalized PageRank, T is set so that teleportation results in equal probabilities of landing at one of the disease-specific seed genes and zero probability of teleporting to any other genes within the genetic network. PageRank evaluation of a network can be thought of as a random walk from one node to another, and the number of times one lands on a specific node decides its final PageRank. Teleportation introduces a random jump from a node to an unconnected node. By limiting teleportation to seed genes, we effectively increase the rank of all seed genes and all genes within the network neighborhood of seed genes. The α value for this calculation was optimized using a heuristic approach, and 0.95 was found to perform best; however, results were stable across most α values (see Supplementary Text online).
Personalized PageRank is calculated with each of the three different A matrices and divided by the corresponding basic PageRanks to obtain a score, or a fold change in the basic versus personalized PageRank score. This division step is required to suppress the effect of highly connected hub genes that would otherwise almost always get a high rank. The filtered genes to be ranked then are sorted from high to low scores and ranked accordingly. The final rank for any gene is the lowest (best) rank it obtains among the set of its three ranks, that is, one for each scale of A.
This ranking methodology effectively integrates seed gene information from multiple sources and places some emphasis on local hub genes versus less-well-connected genes because the ranks depend not only on the distance between nodes but also on the importance of each node and its interacting partners within the network at large.
Symptom ranking case
Although our test cases had a set of defined genes used as seeds to identify a novel gene–disease relationship, in a real-world scenario the most closely related disease may not be obvious. In this case we propose that seed genes can be based on the symptoms of the individual. To simulate this, a recently solved idiopathic disease from the IDIOM trial at the Scripps Translational Science Institute was used as an example. The proband, referred to as ID1, was a 14-year-old girl of European ancestry at the time of enrollment. ID1 initially presented with hypotonia, weakness, and abnormal involuntary movements. She had delayed development and showed paroxysmal movement of limbs, which was enhanced at night.9
A set of ID1’s symptoms was compiled using the physician’s summary. The publicly available online tool Phenomizer (http://compbio.charite.de/phenomizer/)28,29 was then used to identify a list of potential diseases associated with her symptoms. The causative genes underlying diseases identified by Phenomizer were used as seeds in the PIN-rank teleportation matrix, where the weight of each gene corresponds to the −log(P value) of the disease-to-symptoms association. These values are normalized so that teleportation probabilities sum to 1.0. Finally, candidate causative genes identified in the ID1 trio analysis were ranked using the PIN-rank method utilizing these symptom-derived seeds.
Results
Overall ranking results
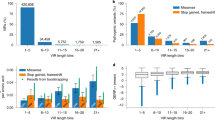
Twenty-two test trios were implanted with 93 test disease-causative genes (Supplementary Table S2 online), for a total of 2,046 simulated family-based disease–gene identification test cases. Each simulated test case was filtered to produce a set of candidate disease-causative genes, as described in Materials and Methods. Each test case resulted in between 4 and 27 candidate disease-causative genes (Supplementary Table S1 online). These, along with the implanted gene. were ranked with PIN-rank, using seeds (Supplementary Table S2 online) for the disease corresponding to the implanted gene to build the teleportation matrix (described in Materials and Methods). The implanted gene was the top-ranked gene in 34.07% of all cases and was present in the top three ranked genes in 55.43% of cases when ranked via PIN-rank, compared with 6.62 and 19.85% by random chance (permutation analysis) ( Figure 1a ). Thus PIN-rank produced a much greater proportion of successes than expected by random ranking (P < 1 × 10−6, Mann–Whitney U test).
Comparison of PIN-rank performance using disease-matched seeds. The cumulative percentage of candidate genes achieving each rank threshold after ranking (x-axis) plotted for different cases. (a) Graph showing PIN-rank with disease-matched seed genes (solid line); PIN-rank with alternative disease seed genes (dotted and dashed line); random ranks (dotted line); and ranking based on path lengths (dashed line). (b) Graph showing PIN-rank with disease-matched seed genes (solid line); PIN-rank with alternative seed genes from the different disease class (dashed line); PIN-rank with alternative seed genes from the same disease class (dotted and dashed line); and random ranking (dotted line). PIN, phenotype informed network.
To confirm that the success in ranking stems from the use of appropriate seed genes rather than some other general characteristic of the disease-causing genes, we selected all test gene–disease associations and randomly swapped the set of seed genes used to rank each test gene–disease association. Each test gene–disease association was then ranked with the seeds from all other test gene–disease associations. Only 9.10% of all test gene–disease association rerankings achieved rank 1, and 27.41% of all test gene–disease association rerankings scored within the top three ranks ( Figure 1a ). This performance is superior to random ranking but inferior to ranking based on exactly matched seeds, confirming that the methodology and selection of appropriate seed genes, rather than some general characteristic of disease-causing genes, drives the performance of our ranking algorithm. The observed improvement in ranking with swapped seed genes versus completely random ranking likely occurred because our implanted genes belong to similar diseases. To test this hypothesis, we compared the performance of PIN-rank when swapping seed genes within or across diseases classes (Supplementary Table S3 online). When using similar diseases, the implanted gene was the top-ranked gene in 14.38% of all cases and was present in the top three ranked genes in 37.19%, compared with 7.27 and 24.04% when ranking with diseases from alternative disease classes ( Figure 1b ). The improved performance relative to random ranking (P < 1 × 10−6, Mann–Whitney U test) suggests that similar diseases identified by symptom matching would be effective for disease gene identification.
Network characteristics by rank
Next, we investigated whether network characteristics influenced the performance of our ranking methodology. First, we determined whether disease–gene connectivity, measured as degree and betweenness centrality, was correlated with ranking performance. Disease–gene degree centrality (P = 0.032; r2 = 0.049) had significant but very low correlation, whereas disease–gene betweenness centrality (P = 0.158; r2 = 0.022) was not significantly correlated with PIN ranking across the 22 genomes. As expected, our methodology requires some minimal level of connectivity information for the disease gene, but the lack of any substantial correlation with gene connectivity suggests no major bias for the detection of hublike disease genes. This ability to detect non-hublike disease genes is important given that a subset of nonessential disease genes show no tendency to act as hubs in genetic networks.30
A common method to identify novel disease-causative genes is to simply prioritize candidate genes closely connected to known disease-causative genes. Therefore, we performed a series of comparisons to demonstrate the superiority of PIN-rank versus a shortest path–based approach to ranking implanted disease-causative genes (CG). As shown in Figure 1a , overall PIN-rank achieved superior results as compared with shortest path–based ranking (P < 1 × 10−6, Mann–Whitney U test). The CG achieved an equivalent or superior rank using the PIN-rank approach as compared with the shortest-path approach in 81.13% of cases.
To explore the nature of this performance advantage in more detail, we introduce a metric denoted as number of paths. This metric represents the number of seed genes required so that the sum of the shortest paths between the seed genes to the CG is less than the sum of the same number of shortest paths between the most highly ranked noncausal candidate gene and its closest seeds. For example, if the CG is the closest candidate gene to any seed gene, then the number of paths metric is equal to 1. If the noncausal candidate gene is the closest candidate gene to a seed gene, then we determine whether the sum of the path lengths between the CG and its two closest seed genes is less than the sum of paths from the noncausal candidate gene to its two closest seed genes; if so, the number of paths metric is equal to 2. This process is continued until all seed genes are exhausted.
Using this metric, we evaluated the performance of the PIN-rank method as compared with the shortest path ranking approach for CGs at different numbers of path values. We observed that the superior performance of PIN-rank was achieved across a range of numbers of path distances between the seed genes and CG ( Figure 2a ). That is, the overall performance of the PIN-rank method is superior regardless of whether the CG is closest to seeds.
PIN-rank performance with respect to path lengths. (a) The number of paths metric, that is, the number of seed genes required such that the sum of paths to those from the disease-causative gene is smaller than the sum of the same number of paths for the top-ranking non-disease-causative gene and its nearest seeds (x-axis) plotted against the median rank observed for PIN ranking method (dots) and the median rank observed for ranking based on path lengths (crosses). (b) Successes, defined as the causative gene achieving the top rank among all candidate genes, or failures observed for each number of paths as a percentage of total successes and failures. PIN, phenotype informed network.
Moreover, although overall performance tends to deteriorate as the distance from the CG to the seed genes increases, the method continues to successfully identify the CG across a range of path length distances. To demonstrate this, we plotted the frequency of successes across different numbers of paths, where success is defined as when the CG is the top-ranked gene. As shown in Figure 2b , in about 20% of successful cases the CG was identifiable not just because it was the closest candidate gene to a seed gene. That is to say, in 20% of our successes, the identification of the novel disease-causative gene required the synergistic contribution of multiple seed genes in order to be correctly identified. Moreover, in ~43% of these successes, after considering all seed genes, the CG was never the closest candidate gene to any subset of seed genes. In these cases the nature of the connectivity of the CG to the seed genes played an important role in causing the CG to be highly ranked.
Thus the successful identification of disease genes via the PIN-rank method is superior to the trivial exercise of connecting candidate genes to seed genes known to cause similar (or the same) diseases. Similarly, nearly all genes in the network are two steps away from a seed gene, yet our method is often able to accurately distinguish between the true CG and other nearby noncausal candidate genes. These successes are due to the integration of information emanating from multiple seeds and the nature of the connectivity from seeds to CGs. The observation that multiple seed genes contribute to the accurate identification of the correct disease-causative gene, coupled with the observation that ranking with seed genes of a different yet related disease leads to the superior ranking relative to random chance, led us to propose that selecting seed genes based purely on the symptoms of an affected case, rather than the somewhat subjective selection of a related disease, may be an effective means of prioritizing disease-causative genes in the case of previously undescribed diseases.
Symptom ranking example
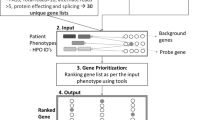
The initial filtering of ID1’s genome resulted in 16 possible causative genes (Supplementary Table S4 online). The Phenomizer output using symptoms compiled from the physician’s summary (Supplementary Table S4 online) had a total of 7,449 diseases with 2,887 total associated genes, of which ~2,300 had a nonzero P value, all of which were used as weighted seeds in the ranking. The top 100 diseases and corresponding genes are presented in Supplementary Table S5 online, with ADCY5, the known causative gene for this condition,9 removed. PIN-rank correctly identified the disease-causative gene as ADCY5, ranking it first among the 16 possible genes. Figure 3 displays the relationship between the top few symptom-derived seed genes and candidate causative genes. Although ADCY5 is connected directly to two seed genes, other candidate genes were similarly connected or, in the case of TGFB2, connected to a larger number of seed genes. Yet the nature of the connection between ADCY5 and the seed genes, especially its central position among two larger clusters of interconnected seed genes, resulted in a superior ranking. This reinforces our earlier conjecture that more distant seed genes play an important role in the ranking of candidate genes and that the PIN-rank approach effectively utilizes these more distal and synergistic gene relationships.
Network view of candidate genes and top seed genes. The network of candidate genes and a few of the top symptom-derived genes (seeds) used for the ranking: dark green indicates candidate disease-causative genes; light green indicates ADCY5; light red represents seed genes; and yellow indicates other closely connected genes. As evident, ADCY5 is connected directly to two seed genes. Although other candidate genes have similar direct connections, ADCY5 has a more central position among two larger clusters of interconnected seed genes, resulting in it obtaining the top rank.
Discussion
It is well known that similar diseases are mediated by genes that are neighbors to one other within a genetic network30 and that similar diseases can be identified through phenotypic similarity.28 Thus, it makes intuitive sense that novel gene–disease relationships can be identified by cross-correlating genetic and phenotypic data. A number of systems for prioritizing disease genes in other contexts, using a systems biology–based approach, have been described.12,13,31,32 In this article we present one such approach to this problem and demonstrate its success by strictly benchmarking performance through temporal separation of the information being predicted from the information used to make the prediction. This is a key and often overlooked component of any realistic performance evaluation.
PHEVOR,31 a similar method, uses functional ontologies rather than genetic networks. Although certainly useful for the phenotype-driven identification of known gene–disease relationships, how well this method would perform on novel gene–disease relationships is not clear. Although the authors attempted to simulate this scenario by removing all known phenotype relationships for the known causative gene, no separation of gene ontology information before and after the discovery of the gene–disease relationship was performed—in contrast to our benchmarks, which temporally separate the genetic network information used to identify novel gene–disease relationships. Although a direct comparison should be done, performing a comparison is not possible without assembling training and testing data, as we have done herein, where knowledge obtained from the discovery being simulated does not bias the training data. Regardless, our simulations indicate superior performance over that reported by PHEVOR and highlights the fact that absolutely no prior knowledge regarding gene function is required to drive PIN-rank, given that genetic networks can be inferred simply through unbiased gene-expression data.33,34
Although we have limited the identification of seed genes based on matching to known human phenotypes through Phenomizer,28 a few opportunities for improvement are apparent. A recently described method, PHIVE,35 uses cross-species phenotypic information to prioritize candidate genes in human sequencing studies. Essentially, the phenotype of the diseased individual under investigation is compared with phenotypes in model organisms caused by known gene defects to identify candidate genes that may explain their phenotype. This process relies on the existence of model organisms with the appropriate candidate gene perturbed. Performance of this method is overestimated because the majority of these mouse models (69%) were generated as models of the disease in question.35 However, our network-based approach to this problem could potentially circumvent this issue by not requiring a gene-specific model of the disease in question.
A few limitations exist for our approach. First, the method will suffer because of the incompleteness of genetic networks. However, the ability to reconstruct genetic networks from unbiased genomic data suggests this is a minor limitation. Second, phenotype information for rare Mendelian diseases can be limited. A number of initiatives have recently been launched to address this limitation. Third, there is a chance that the candidate genome sequence data might not contain the disease-causing variant, due to inaccurate or incomplete variant calling. Finally, our method is not capable of identifying gene–disease relationships mediated through completely novel biological processes or mechanisms of action. In other words, the genetic network seeding process biases the identification of novel gene–disease relationship toward those mediated through biological processes already associated with similar diseases. Gene-disease relationships mediated through completely novel biological processes will be the most difficult molecular diagnoses to make using any analytical model, short of identifying multiple individuals affected with the same phenotype through the same underlying mechanism. Nevertheless, we have demonstrated the power of our approach to identify novel gene–disease relationships in family-based sequencing studies.9
Disclosure
A.T. is a cofounder of Cypher Genomics. P.H.P. is an employee at Cypher Genomics. B.M. declares no conflict of interest.
References
Gratten J, Wray NR, Keller MC, Visscher PM . Large-scale genomics unveils the genetic architecture of psychiatric disorders. Nat Neurosci 2014;17:782–790.
Boycott KM, Vanstone MR, Bulman DE, MacKenzie AE . Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nat Rev Genet 2013;14:681–691.
Cooper DN, Chen JM, Ball EV, et al. Genes, mutations, and human inherited disease at the dawn of the age of personalized genomics. Hum Mutat 2010;31:631–655.
Choi M, Scholl UI, Ji W, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A 2009;106:19096–19101.
Ng SB, Buckingham KJ, Lee C, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet 2010;42:30–35.
Ng SB, Bigham AW, Buckingham KJ, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet 2010;42:790–793.
Ku CS, Naidoo N, Pawitan Y . Revisiting Mendelian disorders through exome sequencing. Hum Genet 2011;129:351–370.
Lyon GJ, Wang K . Identifying disease mutations in genomic medicine settings: current challenges and how to accelerate progress. Genome Med 2012;4:58.
Chen YZ, Friedman JR, Chen DH, et al. Gain-of-function ADCY5 mutations in familial dyskinesia with facial myokymia. Ann Neurol 2014;75:542–549.
Girirajan S, Rosenfeld JA, Coe BP, et al. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N Engl J Med 2012;367:1321–1331.
Dorschner MO, Amendola LM, Turner EH, et al.; National Heart, Lung, and Blood Institute Grand Opportunity Exome Sequencing Project. Actionable, pathogenic incidental findings in 1,000 participants’ exomes. Am J Hum Genet 2013;93:631–640.
Koboldt DC, Larson DE, Sullivan LS, et al. Exome-based mapping and variant prioritization for inherited Mendelian disorders. Am J Hum Genet 2014;94:373–384.
Zemojtel T, Köhler S, Mackenroth L, et al. Effective diagnosis of genetic disease by computational phenotype analysis of the disease-associated genome. Sci Transl Med 2014;6:252ra123.
Abecasis GR, Auton A, Brooks LD, et al.; 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature 2012;491:56–65.
Drmanac R, Sparks AB, Callow MJ, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 2010;327:78–81.
McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–1303.
DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491–498.
Van der Auwera GA, Carneiro MO, Hartl C, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 2013;11:11.10.1–11.10.33.
International HapMap Consortium. The International HapMap Project. Nature 2003;426:789–796.
National Heart, Lung, and Blood Institute (NHLBI). NHLBI Exome Sequencing Project (ESP). NHLBI: Seattle, WA, 2011.
Borrell B . Sequencing projects bring age-old wisdom to genomics. Nat Med 2011;17:1329.
Wellderly Study: uncovering the genetic secrets of good health. http://www.stsiweb.org/translational_research/research_highlights/healthy_aging/. Accessed February 2014.
Page L, Brin S, Motwani R, Winograd T . The PageRank Citation Ranking Bringing Order to the Web. Stanford InfoLab, 1999. http://ilpubs.stanford.edu:8090/422/
Bonacich P, Lloyd P . Eigenvector-like measures of centrality for asymmetric relations. Soc Networks 2001;23:191–201.
Haveliwala T, Kamvar S, Jeh G . An Analytical Comparison of Approaches to Personalizing PageRank. Stanford InfoLab, 2003. http://ilpubs.stanford.edu:8090/cgi/export/422/EndNote/ilprints-eprint-422.enw
Jensen LJ, Kuhn M, Stark M, et al. STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res 2009;37(Database issue):D412–D416.
Csardi G, Nepusz T . The igraph software package for complex network research. InterJournal 2006; Complex Systems: 1695.
Köhler S, Schulz MH, Krawitz P, et al. Clinical diagnostics in human genetics with semantic similarity searches in ontologies. Am J Hum Genet 2009;85:457–464.
Köhler S, Doelken SC, Mungall CJ, et al. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res 2014;42(Database issue):D966–D974.
Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabási AL . The human disease network. Proc Natl Acad Sci U S A 2007;104:8685–8690.
Singleton MV, Guthery SL, Voelkerding KV, et al. Phevor combines multiple biomedical ontologies for accurate identification of disease-causing alleles in single individuals and small nuclear families. Am J Hum Genet 2014;94:599–610.
Javed A, Agrawal S, Ng PC . Phen-Gen: combining phenotype and genotype to analyze rare disorders. Nat Methods 2014;11:935–937.
Margolin AA, Nemenman I, Basso K, et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics 2006;7(suppl 1):S7.
Zhang B, Horvath S . A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 2005;4:Article17.
Robinson PN, Köhler S, Oellrich A, et al.; Sanger Mouse Genetics Project. Improved exome prioritization of disease genes through cross-species phenotype comparison. Genome Res 2014;24:340–348.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Text
(DOC 154 kb)
Supplementary Tables
(XLS 93 kb)
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Molparia, B., Pham, P. & Torkamani, A. Symptom-driven idiopathic disease gene identification. Genet Med 17, 859–865 (2015). https://doi.org/10.1038/gim.2014.202
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2014.202