Abstract
Hepatic steatosis is common in obese individuals with hyperinsulinemia and is an important hepatic manifestation of metabolic syndrome. Sterol regulatory binding protein-1c (SREBP-1c) is a master regulator of lipogenic gene expression in the liver. Hyperinsulinemia induces transcription of SREBP-1c via activation of liver X receptor (LXR) and specificity protein 1 (Sp1). Cilostazol is an antiplatelet agent that prevents atherosclerosis and decreases serum triglyceride levels. However, little is known about the effects of cilostazol on hepatic lipogenesis. Here, we examined the role of cilostazol in the regulation of SREBP-1c transcription in the liver. The effects of cilostazol on the expression of SREBP-1c and its target genes in response to insulin or an LXR agonist (T0901317) were examined using real-time RT-PCR and western blot analysis on cultured hepatocytes. To investigate the effect of cilostazol on SREBP-1c at the transcriptional level, transient transfection reporter assays and electrophoretic mobility shift assays (EMSAs) were performed. Cilostazol inhibited insulin-induced and LXR-agonist-induced expression of SREBP-1c and its downstream targets, acetyl-CoA carboxylase and fatty acid synthase, in cultured hepatocytes. Cilostazol also inhibited activation of the SREBP-1c promoter by insulin, T0901317 and Sp1 in a luciferase reporter assay. EMSA analysis showed that cilostazol inhibits SREBP-1c expression by repressing the binding of LXR and Sp1 to the promoter region. These results indicate that cilostazol inhibits insulin-induced hepatic SREBP-1c expression via the inhibition of LXR and Sp1 activity and that cilostazol is a negative regulator of hepatic lipogenesis.
Similar content being viewed by others
Introduction
Hepatic steatosis is common in obese individuals with insulin resistance and is an important hepatic manifestation of metabolic syndrome.1, 2 Hyperinsulinemia caused by peripheral insulin resistance potently induces the expression of enzymes that catalyze de novo synthesis of fatty acids in the liver.3 The ability of insulin to activate lipogenesis is mediated by transcriptional activation of sterol regulatory binding protein-1c (SREBP-1c).4 SREBP-1c is a membrane-bound transcription factor of the basic helix-loop-helix leucine zipper family that regulates the expression of the key enzymes implicated in hepatic lipid metabolism. The role of SREBP-1c in fatty liver development in obese individuals with insulin resistance is well established. Regulation of hepatic SREBP-1c expression is largely dependent on hormonal stimuli.5 Insulin increases SREBP-1c expression by promoting the cleavage of the SREBP-1c precursor into its active nuclear form and by activating liver X receptor (LXR) and specificity protein 1 (Sp1).6, 7, 8 Therefore, identification of compounds that inhibit these transcription factors may provide effective treatment options for hepatic steatosis.
The phosphodiesterase III inhibitor cilostazol is an antiplatelet agent used to treat patients with diabetic vascular complications.9, 10 Several lines of evidence have suggested that cilostazol is also an effective treatment for dyslipidemia.11, 12, 13 Antiplatelet agents, particularly cilostazol, attenuate hepatic steatosis, inflammation and fibrosis in vivo via the suppression of oxidative stress-induced and platelet-derived growth factor-induced mitogen-activated protein kinase activation.14 Moreover, clinical trials have shown that, in addition to its ability to reduce the progression of carotid intima-media thickness, cilostazol also decreases serum triglyceride and low-density lipoprotein cholesterol levels and increases high-density lipoprotein cholesterol levels.11 Taken together, results from both preclinical and clinical studies suggest that cilostazol may be used to inhibit hepatic steatosis; however, little is currently known about the effect of cilostazol on hepatic lipogenesis and the mechanism by which cilostazol improves lipid dysfunction. In this study, we examined the inhibitory role of cilostazol on insulin-stimulated SREBP-1c expression.
Materials and methods
Chemical and antibodies
Cilostazol was donated by the Otsuka Pharmaceutical Co. (Otsuka, Japan). Insulin was provided by Novo Nordisk (Bagsvaerd, Denmark). Radiochemicals ([γ-32P]ATP) were purchased from Perkin Elmer (Boston, MA, USA). T0901317 was from Sigma (St Louis, MO, USA). The SREBP-1 antibody was purchased from BD Biosciences (San Jose, CA, USA) and the β-actin antibody was purchased from Sigma. The goat anti-mouse secondary antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Plasmids
Luciferase reporter constructs containing the wild-type SREBP-1c promoter (pSREBP(−1516/+40)-luciferase) and a mutant SREBP-1c promoter containing two LXR-binding site mutations (pSREBP(−1516/+40) mutLXR1/mutLXR2-luciferase) were kind gifts from Dr Cagen (The University of Tennessee, Knoxville, TN, USA). A synthetic Sp1 promoter–reporter construct containing four Sp1-binding sites (GC boxes) ([SP1]X4/luc) and the pCMV-Sp1 expression plasmid were kindly provided by Dr Chang (Catholic University of Daegu, Korea).
Cell culture
The HepG2 human hepatoma cell line and the H4IIE rat hepatoma cell line were purchased from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured at 37 °C and 5% CO2 in Minimum Essential Medium (Gibco BRL/Life Technologies, Grand Island, NY, USA) containing L-glutamine. The culture medium was supplemented with 10% fetal bovine serum, 1 mM sodium pyruvate, 1.5 g l−1 sodium bicarbonate and antibiotics. Equal numbers of cells were seeded onto tissue culture plates and cells were rendered quiescent by incubation for 24 h in growth medium containing 0.5% fetal bovine serum.
Primary cultures of hepatocytes
C57BL/6 mouse hepatocytes were isolated by perfusing the liver via the portal vein. The liver was first perfused with resuspension buffer (140 mM NaCl, 5.4 mM KCl, 0.44 mM KH2PO4, 0.34 mM Na2HPO4, 0.5 mM EGTA and 25 mM Tricine; pH 7.2) at 4 ml min−1 for 10 min, and then perfused with collagenase solution (9.5 g l−1 HBSS, 0.8 mM NaHCO3, 0.002% DNase I, 0.7 g l−1 collagenase type I (Worthington Biochemical Corporation, Lakewood, NJ, USA); pH 7.2), at 4 ml min−1 for 10 min. After perfusion, the liver tissue was shaken for 15 min at 37 °C and passed through a mesh filter (85 μm nylon mesh). Hepatocytes were collected by centrifugation at 500 r.p.m. for 5 min at 4 °C, resuspended in William’s E medium (Sigma) and seeded onto collagen type I-coated 60-mm dishes (IWAKI Scitech Kiv., Tokyo, Japan) at a density of 5 × 105 cells ml−1. The viability of hepatocytes, as measured by trypan blue dye exclusion, was routinely >85%. After a 2- to 3-h incubation, the medium was changed to Dulbecco’s Modified Eagle Medium. Hepatocytes were used subsequently for quantitative real-time RT-PCR.
Real-time PCR
Total RNA was isolated from cells using Trizol reagent (Life Technologies), according to the manufacturer’s instructions. Reverse transcription was performed using a first-strand cDNA synthesis kit (Fermentas, Hanover, MD, USA). Quantitative real-time RT-PCR was performed using a SYBR Green PCR Master Mix Kit (TOYOBO, Osaka, Japan) and a Light Cycler 480 instrument (Roche Diagnostics, Indianapolis, IN, USA). PCR parameters were as follows: 45 cycles of 95 °C for 30 s, 60 °C for 10 s and 72 °C for 15 s. Primer sequences were as follows: rat SREBP-1 5′-CCATCGACTACATCCGCTTCTT-3′ (forward) and 5′-ACTTCGCAGGGTCAGGTTCTC-3′ (reverse), rat acetyl-CoA carboxylase (ACC) 5′-CAGGTGCTCAAGTTTGGTGC-3′ (forward) and 5′-TGGGTCGATCACAACCCAAG-3′ (reverse), and rat fatty acid synthase (FAS) 5′-GGACTTGGGTGCCGATTA-3′ (forward) and 5′-GGTCGTGGCTTCATGGTAGG-3′ (reverse).
Immunoblot analysis
Cells were washed twice with phosphate-buffered saline and then suspended in radioimmunoprecipitation assay buffer containing 50 mM Tris (pH 7.4), 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 0.25% sodium deoxycholate, 1 mM phenylmethysulfonyl fluoride, 1 mg ml−1 leupeptin, 1 mg ml−1 aprotinin, 1 mM sodium orthovanadate and 1 mM sodium fluoride. Cells were lysed on ice for 30 min and the cell lysate was collected after centrifugation at 12 000 r.p.m. for 10 min. Protein quantification was performed by Bio-Rad Protein Assay (Bio-Rad, Richmond, CA, USA). Cell lysates were electrophoresed using SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Millipore Corporation, Bedford, MA, USA). The membrane was blocked using skim milk buffer. After blocking, the membrane was incubated with an anti-SREBP-1 monoclonal antibody (1:500 dilution) and then washed. Antibody binding was detected using a horseradish peroxidase-linked secondary antibody (1:2500 dilution) and an ECL western blotting detection system (Amersham, Buckinghamshire, UK). The membrane was reblotted with an anti-actin antibody (1:5000 dilution) to verify equal protein loading in each lane. Densitometric measurements of the bands were performed using the UN-SCAN-IT software (Silk Scientific Corporation, Orem, UT, USA).
In vivo transfection and reporter gene assays
HepG2 cells were plated onto 12-well plates at a density of 1 × 105 cells per well and then cultured for 1 day. Cells were transiently transfected with promoter–reporter constructs using Lipofectamine 2000 regent (Invitrogen, Grand Island, NY, USA). Cells were co-transfected with a luciferase promoter–reporter construct and an expression plasmid containing β-galactosidase, as an internal control. Transfection was allowed to proceed for 5 h and then the cells were washed to remove unincorporated plasmid. Cells were harvested ∼24 h after transfection and luciferase, and β-galactosidase activity in 20 μl of cell lysate was measured. Luciferase activity was read using a Sirius luminometer (Titertek-Berthold, Pforzheim, Germany) and a luciferase assay system (Promega, Madison, WI, USA), according to the manufacturer’s instructions. Luciferase activity was normalized to β-galactosidase activity.
Electrophoretic mobility shift assay
Nuclear extracts were isolated from HepG2 cells using the NucBuster protein extraction kit (Calbiochem, La Jolla, CA), according to the manufacturer’s instructions. After centrifuging cells at 12 000 r.p.m. for 5 min, supernatants containing the nuclear extracts were collected and quantified using a protein assay kit (Bio-Rad). Nuclear extracts (6 μg) were incubated with 60 000 c.p.m. of a 32P-labeled oligomer containing the LXR-binding site (5′-CAGTGACCGCCAGTAACCCCAGC-3′) or the Sp1-binding site (5′-TAACGGGGCGGGGCG AAT-3′), as a probe. Competition experiments were performed with unlabeled oligomer containing the LXR- or Sp1-binding site. Supershift experiments were performed by incubating the binding reaction with an anti-LXR antibody. Samples were loaded onto 4% native polyacrylamide gels and electrophoresed in 0.5% Tris-borate-EDTA buffer at 150 V for 2 h. Gels were then dried and visualized using autoradiography.
Statistical analysis
Data were evaluated using analysis of variance followed by Fisher’s least significant difference post-hoc test. All data are expressed as the mean±s.e.m. A P-value of <0.05 was considered statistically significant. All experiments were performed at least three times.
Results
Cilostazol inhibits insulin-stimulated expression of SREBP-1c
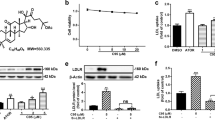
The effect of cilostazol on insulin-stimulated expression of SREBP-1c mRNA and protein was examined in H4IIE cells. Treatment of cells with 100 nM insulin increased the expression of SREBP-1c mRNA (Figure 1a), as well as precursor and nuclear SREBP-1 protein (Figure 1c); however, these responses were inhibited by co-treatment of cells with 70 μM cilostazol (Figures 1a and c). Cilostazol treatment also inhibited insulin-stimulated expression of the two SREBP-1c target genes ACC and FAS (Figure 1a). The inhibitory effect of cilostazol on insulin-stimulated SREBP-1c and SREBP-1c target genes was also observed in mouse primary hepatocytes (Figure 1b). Transient transfection of HepG2 cells with the human SREBP-1c promoter–reporter construct showed that cilostazol inhibited insulin-stimulated SREBP-1c mRNA expression at the level of transcription in a dose-dependent manner (Figure 1d).
The effect of cilostazol on insulin-stimulated sterol regulatory binding protein-1c (SREBP-1c) expression. (a) Real-time RT-PCR analysis of SREBP-1c, acetyl-CoA carboxylase (ACC) and fatty acid synthase (FAS) mRNA levels in H4IIE cells. Expression of GAPDH mRNA was used as an internal control. Data are shown as the mean±s.e.m. of n=3 independent measurements. *P<0.01, as compared with the control. #P<0.05 and ##P<0.01, compared with insulin alone. (b) Real-time RT-PCR analysis of SREBP-1c, ACC and FAS mRNA levels in mouse primary hepatocytes. Expression of GAPDH mRNA was used as an internal control. Data are shown as the mean±s.e.m. of n=3 independent measurements. *P<0.01 and **P<0.001, compared with the control. #P<0.01, compared with insulin alone. (c) Immunoblot analysis of the effect of cilostazol on insulin-stimulated SREBP-1 protein expression. H4IIE cells were pretreated with the indicated concentrations of cilostazol for 24 h and then incubated with insulin (100 nM) for 6 h. Data in the bar graph are the means±s.e.m. of three independent measurements. *P<0.001, compared with the control; #P<0.001, compared with insulin alone. (d) Effect of cilostazol on insulin-stimulated SREBP-1c promoter activity in HepG2 cells. Cells were pretreated with the indicated concentrations of cilostazol for 24 h and then treated with insulin (100 nM) for 6 h. Data are shown as the mean±s.e.m. of n=3 independent measurements *P<0.01, compared with the control. #P<0.01 and ##P<0.001, compared with insulin alone.
Cilostazol inhibits LXR-stimulated expression of SREBP-1c
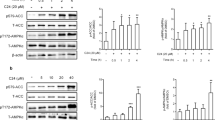
As LXR is an important mediator of insulin-stimulated SREBP-1c expression,15 we examined the ability of cilostazol to inhibit the expression of SREBP-1c induced by the LXR ligand T0901317. Treatment of H4IIE cells with cilostazol significantly inhibited T0901317-stimulated expression of SREBP-1c mRNA (Figure 2a) and protein (Figure 2c). Cilostazol also inhibited T0901317-stimulated expression of ACC and FAS mRNA (Figure 2a). In addition, cilostazol inhibited T0901317-stimulated expression of SREBP-1c, ACC and FAS mRNA in mouse primary hepatocytes (Figure 2b). T0901317 induced a marked increase in SREBP-1c promoter activity, which was inhibited by cilostazol treatment (Figure 3a). Moreover, electrophoretic mobility shift assay (EMSA) analysis showed that cilostazol treatment reduced the T0901317-induced increase in the DNA-binding activity of LXR in a dose-dependent manner (Figure 3b).
The effect of cilostazol on T0901317-stimulated SREBP-1c expression. (a) Real-time RT-PCR analysis of SREBP-1c, acetyl-CoA carboxylase (ACC), and fatty acid synthase (FAS) mRNA levels in H4IIE cells. Expression of GAPDH mRNA was used as an internal control. Data are shown as the mean±s.e.m. of n=3 independent measurements. *P<0.01 and **P<0.001, compared with the control; #P<0.001, compared with T0901317 alone. (b) Real-time RT-PCR analysis of SREBP-1c, ACC and FAS mRNA levels in mouse primary hepatocytes. Expression of GAPDH mRNA was used as an internal control. Data are shown as the mean±s.e.m. of n=3 independent measurements. *P<0.01 and **P<0.001, compared with the control; #P<0.01, compared with T0901317 alone. (c) Immunoblot analysis of the effect of cilostazol on T0901317-stimulated SREBP-1c protein expression. H4IIE cells were incubated with T0901317 (1 μM) for 24 h, with the indicated concentrations of cilostazol. Data in the bar graph are the means±s.e.m. of three independent measurements. *P<0.01, compared with the control; #P<0.01 and ##P<0.001, compared with T0901317 alone.
The effect of cilostazol on T0901317-induced sterol regulatory binding protein-1c (SREBP-1c) promoter activity and liver X receptor (LXR)-binding activity. (a) The effect of cilostazol on T0901317-stimulated SREBP-1c promoter activity in HepG2 cells. Cells were pretreated with the indicated concentration of cilostazol for 24 h and then treated with T0901317 (1 μM) for a further 24 h. Data are shown as the mean±s.e.m. of n=3 independent measurements.*P<0.01, compared with the control; #P<0.001, compared with T0901317 alone. (b) Electrophoretic mobility shift assay (EMSA) analysis of the effect of cilostazol on the T0901317-induced DNA-binding activity of LXR. HepG2 cells were incubated with T0901317 (1 μM) for 24 h, with the indicated concentrations of cilostazol. Competition (Com) experiments were performed with a 10 × concentration of the unlabeled LXR-binding site oligomer. Supershift (S.S) experiments were performed by incubating the binding reaction with an anti-LXR antibody. Data are shown as the mean±s.e.m. of n=3 independent measurements. *P<0.01, compared with the control; #P<0.001, compared with T0901317 alone.
Cilostazol inhibits insulin-stimulated Sp1 activity
Treatment of transfected cells with cilostazol inhibited the basal (Figure 4a) and the insulin-stimulated (Figure 4b) reporter activity of the LXR-mut/SREBP-1c/luc promoter, in which two LXR-binding sites are mutated (Figure 4a), suggesting that cilostazol inhibits transcription factors other than LXR. As Sp1 is known to have an important role in insulin-stimulated SREBP-1c transcription,16 the effect of cilostazol on the Sp1-induced SREBP-1c promoter activity was assessed. As shown in Figure 5a, treatment with cilostazol inhibited Sp1-induced SREBP-1c promoter activity. Moreover, the ability of cilostazol to inhibit insulin-stimulated Sp1 transcriptional activity was examined using a promoter–reporter construct containing four synthetic Sp1-response elements. Cilostazol inhibited the activity of the synthetic Sp1 promoter (Figure 5b). As expected, the combination of Sp1 and insulin further enhanced the activity of the Sp1 promoter; however, cilostazol was still able to inhibit this response in a dose-dependent manner (Figure 5c). Moreover, EMSA analysis showed that cilostazol decreased the DNA-binding activity of Sp1 (Figure 5d).
The effect of cilostazol on the activity of the sterol regulatory binding protein-1c (SREBP-1c) promoter in HepG2 cells. (a) The effect of cilostazol on the activity of a mutant SREBP-1c promoter in which the two liver X receptor (LXR)-binding sites were disrupted, in HepG2 cells. Cells were transfected with the LXR-mut/SREBP-1c/luc promoter–reporter construct and then treated with the indicated concentrations of cilostazol for 24 h. Data are shown as the mean±s.e.m. of n=3 independent measurements. *P<0.05 and **P<0.001, compared with the control. (b) The effect of cilostazol on insulin-induced activity of the LXR-mut/SREBP-1c/luc promoter. HepG2 cells were transfected with the wild-type SREBP-1c promoter (WT/SREBP-1c/luc) or the equivalent promoter region containing disrupted LXR-binding sites (LXR-mut/SREBP-1c/luc), pretreated with the indicated concentrations of cilostazol for 24 h and then stimulated with insulin (100 nM) for 6 h. Data show the mean±s.e.m. of n=3 independent measurements.*P<0.01, compared with the control; **P<0.01, compared with the insulin-induced promoter activity of wild-type SREBP-1c. #P<0.05 and ##P<0.01, compared with the insulin-induced activity of the mutant SREBP-1c promoter.
The effect of cilostazol on specificity protein 1 (Sp1) activity. (a) The effect of cilostazol on Sp1-induced Sterol regulatory binding protein-1c (SREBP-1c) promoter activity. HepG2 cells were co-transfected with the SREBP-1c promoter construct (200 ng per well) and a Sp1 expression construct (100 ng), and then cells were exposed to the indicated concentrations of cilostazol for 24 h. Data represent the means±s.e.m. of n=3 independent measurements. *P<0.01, compared with the control, #P<0.01, compared with Sp1 alone. (b) The effect of cilostazol on Sp1 promoter activity. HepG2 cells were co-transfected with a plasmid containing Sp1 (pCMV-Sp1) and a plasmid containing four synthetic Sp1-response elements ([Sp1]X4luc), and the cells were exposed to the indicated concentrations of cilostazol for 24 h. Data are shown to be the mean±s.e.m. of n=3 independent measurements. *P<0.05, compared with the control. #P<0.05 and ##P<0.01, compared with the Sp1-stimulated [Sp1]X4 promoter activity. (c) The effect of insulin and cilostazol on Sp1 promoter activity. HepG2 cells were co-transfected with pCMV-Sp1 and [Sp1]X4luc, pretreated with the indicated concentrations of cilostazol for 24 h, and then stimulated with insulin (100 nM) for 6 h. Data are shown to be the mean±s.e.m. of n=3 independent measurements. *P<0.05 and **P<0.01, compared with the control. #P<0.001, compared with Sp1 and the insulin-stimulated [Sp1]X4 promoter activity alone. (d) The effect of cilostazol on the insulin-stimulated DNA-binding activity of Sp1. HepG2 cells were incubated with insulin (100 nM) for 6 h, following pretreatment with the indicated concentrations of cilostazol for 24 h. Competition (Com) experiments were performed with a 10 × concentration of the unlabeled Sp1-binding site oligomer. Data are shown as the mean±s.e.m. of n=3 independent measurements. *P<0.01, compared with the control. #P<0.01 and ##P<0.001 as compared with insulin alone.
Discussion
This study demonstrates that cilostazol effectively inhibits insulin-induced SREBP-1c expression in cultured hepatocytes. The results of promoter–reporter assays and EMSA analysis suggest that this repression occurs via the inhibition of the DNA-binding activity of LXR and Sp1.
A recent clinical study showed that cilostazol is an effective treatment for dyslipidemia and is particularly effective at reducing plasma triglyceride levels.11 Cilostazol reportedly reduces plasma triglycerides via an increase in lipoprotein lipase activity;17 however, the exact molecular mechanism through which cilostazol lowers triglycerides is still unclear. Increased plasma triglyceride levels are a common characteristic of the dyslipidemia associated with insulin resistance, and an increase in de novo hepatic lipogenesis is attributed to hypertriglyceridemia in the insulin-resistant state.18 Previous studies have demonstrated that SREBP-1c is a master regulator of hepatic lipogenesis.19, 20, 21 Hyperinsulinemia caused by insulin resistance stimulates hepatic SREBP-1c, resulting in fat synthesis and hepatic steatosis.22 In this study, treatment of cultured hepatocytes with insulin increased the transcription of SREBP-1c and expression of its target lipogenic genes, ACC and FAS; these responses were effectively inhibited by treatment of cells with cilostazol. To determine whether cilostazol regulates SREBP-1c in insulin-independent pathways, the effect of cilostazol on SREBP-1c expression was assessed in the absence of insulin. The results showed that cilostazol modestly decreased basal SREBP-1c expression, demonstrating that cilostazol has insulin-independent activity (data not shown). Therefore, our results support the recent clinical data showing that cilostazol improves the lipid profile by reducing serum triacylglycerol levels11 and suggest that this may occur via the inhibition of the insulin-stimulated SREBP-1c pathway.
Expression of SREBP-1c is regulated at the transcriptional level by various nutritional and hormonal factors.23 Accumulating evidence has shown that LXR is a potent inducer of insulin-stimulated SREBP-1c transcription.7, 20, 24 In this study, EMSA analysis and promoter–reporter assays showed that cilostazol attenuates LXR ligand-stimulated SREBP-1c expression by inhibiting the binding of LXR to the SREBP-1c promoter. In addition, insulin stimulated the luciferase activity of the LXR-mut/SREBP-1c/luc promoter–reporter construct, in which two LXR-binding sites are mutated, and cilostazol inhibited this activity. These results suggest that transcription factors other than LXR are involved in insulin-stimulated SREBP-1c expression. Sp1 also has an important role in insulin-stimulated SREBP-1c transcription.16 Indeed, cilostazol decreased SREBP-1c transcription by modulating the ability of Sp1 to bind to the SREBP-1c promoter. Therefore, it is plausible that the effect of cilostazol on hepatic SREBP-1c expression is multifactorial, occurring via both LXR and Sp1.
In conclusion, this study showed that cilostazol reduces insulin-induced and LXR-agonist-induced expression of SREBP-1c by inhibiting the activity of the LXR and Sp1 transcription factors. These results suggest that the effect of cilostazol on hepatic lipogenesis is partly related to the inhibition of SREBP-1c transcription. Further investigation is required to explore the potentially promising role of cilostazol in the treatment of hepatic steatosis.
References
Targher G . Non-alcoholic fatty liver disease, the metabolic syndrome and the risk of cardiovascular disease: the plot thickens. Diabet Med 2007; 24: 1–6.
Marceau P, Biron S, Hould FS, Marceau S, Simard S, Thung SN et al. Liver pathology and the metabolic syndrome X in severe obesity. J Clin Endocrinol Metab 1999; 84: 1513–1517.
Postic C, Girard J . Contribution of de novo fatty acid synthesis to hepatic steatosis and insulin resistance: lessons from genetically engineered mice. J Clin Invest 2008; 118: 829–838.
Azzout-Marniche D, Becard D, Guichard C, Foretz M, Ferre P, Foufelle F . Insulin effects on sterol regulatory-element-binding protein-1c (SREBP-1c) transcriptional activity in rat hepatocytes. Biochem J 2000; 350 (Pt 2): 389–393.
Kim JB, Sarraf P, Wright M, Yao KM, Mueller E, Solanes G et al. Nutritional and insulin regulation of fatty acid synthetase and leptin gene expression through ADD1/SREBP1. J Clin Invest 1998; 101: 1–9.
Cagen LM, Deng X, Wilcox HG, Park EA, Raghow R, Elam MB . Insulin activates the rat sterol-regulatory-element-binding protein 1c (SREBP-1c) promoter through the combinatorial actions of SREBP, LXR, Sp-1 and NF-Y cis-acting elements. Biochem J 2005; 385: 207–216.
Chen G, Liang G, Ou J, Goldstein JL, Brown MS . Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver. Proc Natl Acad Sci USA 2004; 101: 11245–11250.
Schweizer M, Roder K, Zhang L, Wolf SS . Transcription factors acting on the promoter of the rat fatty acid synthase gene. Biochem Soc Trans 2002; 30: 1070–1072.
Shinoda-Tagawa T, Yamasaki Y, Yoshida S, Kajimoto Y, Tsujino T, Hakui N et al. A phosphodiesterase inhibitor, cilostazol, prevents the onset of silent brain infarction in Japanese subjects with type II diabetes. Diabetologia 2002; 45: 188–194.
Shinohara Y, Gotoh F, Tohgi H, Hirai S, Terashi A, Fukuuchi Y et al. Antiplatelet cilostazol is beneficial in diabetic and/or hypertensive ischemic stroke patients. Subgroup analysis of the cilostazol stroke prevention study. Cerebrovasc Dis 2008; 26: 63–70.
Katakami N, Kim YS, Kawamori R, Yamasaki Y . The phosphodiesterase inhibitor cilostazol induces regression of carotid atherosclerosis in subjects with type 2 diabetes mellitus: principal results of the Diabetic Atherosclerosis Prevention by Cilostazol (DAPC) study: a randomized trial. Circulation 2010; 121: 2584–2591.
Nakaya K, Ayaori M, Uto-Kondo H, Hisada T, Ogura M, Yakushiji E et al. Cilostazol enhances macrophage reverse cholesterol transport in vitro and in vivo. Atherosclerosis 2010; 213: 135–141.
Nakaya Y, Minami A, Sakamoto S, Niwa Y, Ohnaka M, Harada N et al. Cilostazol, a phosphodiesterase inhibitor, improves insulin sensitivity in the Otsuka Long-Evans Tokushima Fatty Rat, a model of spontaneous NIDDM. Diabetes Obes Metab 1999; 1: 37–41.
Fujita K, Nozaki Y, Wada K, Yoneda M, Endo H, Takahashi H et al. Effectiveness of antiplatelet drugs against experimental non-alcoholic fatty liver disease. Gut 2008; 57: 1583–1591.
Dif N, Euthine V, Gonnet E, Laville M, Vidal H, Lefai E . Insulin activates human sterol-regulatory-element-binding protein-1c (SREBP-1c) promoter through SRE motifs. Biochem J 2006; 400: 179–188.
Deng X, Yellaturu C, Cagen L, Wilcox HG, Park EA, Raghow R et al. Expression of the rat sterol regulatory element-binding protein-1c gene in response to insulin is mediated by increased transactivating capacity of specificity protein 1 (Sp1). J Biol Chem 2007; 282: 17517–17529.
Tani T, Uehara K, Sudo T, Marukawa K, Yasuda Y, Kimura Y . Cilostazol, a selective type III phosphodiesterase inhibitor, decreases triglyceride and increases HDL cholesterol levels by increasing lipoprotein lipase activity in rats. Atherosclerosis 2000; 152: 299–305.
Ginsberg HN, Zhang YL, Hernandez-Ono A . Regulation of plasma triglycerides in insulin resistance and diabetes. Arch Med Res 2005; 36: 232–240.
Becard D, Hainault I, Azzout-Marniche D, Bertry-Coussot L, Ferre P, Foufelle F . Adenovirus-mediated overexpression of sterol regulatory element binding protein-1c mimics insulin effects on hepatic gene expression and glucose homeostasis in diabetic mice. Diabetes 2001; 50: 2425–2430.
Liang G, Yang J, Horton JD, Hammer RE, Goldstein JL, Brown MS . Diminished hepatic response to fasting/refeeding and liver X receptor agonists in mice with selective deficiency of sterol regulatory element-binding protein-1c. J Biol Chem 2002; 277: 9520–9528.
Shimano H, Yahagi N, Amemiya-Kudo M, Hasty AH, Osuga J, Tamura Y et al. Sterol regulatory element-binding protein-1 as a key transcription factor for nutritional induction of lipogenic enzyme genes. J Biol Chem 1999; 274: 35832–35839.
Browning JD, Horton JD . Molecular mediators of hepatic steatosis and liver injury. J Clin Invest 2004; 114: 147–152.
Osborne TF . Sterol regulatory element-binding proteins (SREBPs): key regulators of nutritional homeostasis and insulin action. J Biol Chem 2000; 275: 32379–32382.
Schultz JR, Tu H, Luk A, Repa JJ, Medina JC, Li L et al. Role of LXRs in control of lipogenesis. Genes Dev 2000; 14: 2831–2838.
Acknowledgements
This work was supported by grants from the National Research Foundation (2012R1A2A2A01043867, Future-Based Technology Development Program 2010-0019514); the Basic Science Research Program (2012R1A1A1010047) of the National Research Foundation of Korea (NRF), which is funded by the Ministry of Education, Science, and Technology; the Korea Health Technology R&D Project of the Ministry of Health & Welfare, Republic of Korea (A111345); and the Korean Diabetes Association Grant (2011).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Jung, YA., Kim, H., Bae, KH. et al. Cilostazol inhibits insulin-stimulated expression of sterol regulatory binding protein-1c via inhibition of LXR and Sp1. Exp Mol Med 46, e73 (2014). https://doi.org/10.1038/emm.2013.143
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/emm.2013.143