Abstract
Genome-wide association studies (GWAS) have contributed significantly to the understanding of complex disease genetics. However, GWAS only report association signals and do not necessarily identify culprit genes. As most signals occur in non-coding regions of the genome, it is often challenging to assign genomic variants to the underlying causal mechanism(s). Topologically associating domains (TADs) are primarily cell-type-independent genomic regions that define interactome boundaries and can aid in the designation of limits within which an association most likely impacts gene function. We describe and validate a computational method that uses the genic content of TADs to prioritize candidate genes. Our method, called 'TAD_Pathways', performs a Gene Ontology (GO) analysis over genes that reside within TAD boundaries corresponding to GWAS signals for a given trait or disease. Applying our pipeline to the bone mineral density (BMD) GWAS catalog, we identify ‘Skeletal System Development’ (Benjamini–Hochberg adjusted P=1.02x10−5) as the top-ranked pathway. In many cases, our method implicated a gene other than the nearest gene. Our molecular experiments describe a novel example: ACP2, implicated near the canonical ‘ARHGAP1’ locus. We found ACP2 to be an important regulator of osteoblast metabolism, whereas ARHGAP1 was not supported. Our results via BMD, for example, demonstrate how basic principles of three-dimensional genome organization can define biologically informed association windows.
Similar content being viewed by others
Introduction
Genome-wide association studies (GWAS) have discovered several important disease associations.1 Assigning signals to causal genes is difficult because these signals fall principally within non-coding regions and do not necessarily implicate the nearest gene.2 For example, a signal found in an FTO intron has been shown to physically interact with and lead to differential expression of other genes, but not FTO itself.3 Moreover, evidence suggests that a type 2 diabetes (T2D) GWAS signal at TCF7L2 also influences ACSL5.4
Chromatin interaction studies have discovered genome organization principles including topologically associating domains (TADs).5 TADs are genomic regions defined by increased contact frequency, consistency across cell types and enrichment of insulator element flanks.6 Therefore, TADs can be used as boundaries of where non-coding causal variants will most likely impact tissue-independent function.
The paper is structured in the following manner: First, we present our novel computational method, called TAD_Pathways, which uses TADs to determine candidate genes. Next, we apply our method to bone mineral density (BMD) GWAS findings and test two candidates’ importance in osteoblast function. Our pipeline identified ACP2 as a novel regulator of osteoblast metabolism. A full description of the method and validation is available in the Supplementary Video.
Methods
Computational procedures to identify candidate genes
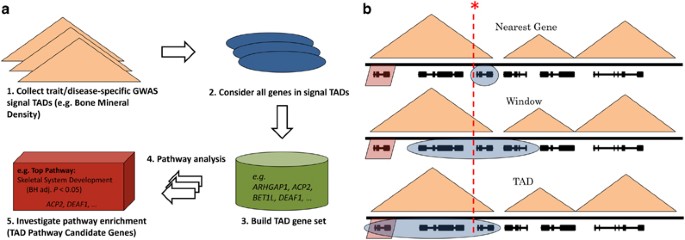
TAD_Pathways is a computational method using publicly available TAD boundaries to prioritize candidate genes from GWAS SNPs (Figure 1a). Alternative approaches assign SNPs to genes based on nearest gene or by an arbitrary or a linkage disequilibrium (LD)-based window of several kilobases (Figure 1b). For full computational methods, refer to the Supplementary Information.
Concepts motivating our approach. TADs are shown as orange triangles, genes are shown as black lines and a genome-wide significant GWAS signal is shown as a dotted red line. (a) The TAD_Pathways method. An example using BMD GWAS signals is shown. (b) Three hypothetical examples are illustrated by a cartoon. The ground truth causal gene is shaded in red. The method-specific selected genes are shaded in blue. The top panel describes a nearest-gene approach. The nearest gene in this scenario is not the gene actually impacted by the GWAS SNP. The middle panel describes a window approach. Based either on linkage disequilibrium or an arbitrarily sized window, the scenario does not capture the true gene. The bottom panel describes the TAD_Pathways approach. In this scenario, the causal gene is selected for downstream assessment.
Here, we use human embryonic stem cell TAD boundaries as reported by Dixon et al.6 and converted to hg19 by Ho et al.7 to build TAD-based gene sets that consists of all genes falling inside TADs implicated with BMD associations. We perform a pathway overrepresentation test8 for the input TAD genes against GO terms.9 This determines if the gene set is associated with any term at a higher probability than by chance. We included both experimentally confirmed and computationally inferred genes, which permit the inclusion of putative genes that do not necessarily have literature support. For validation, we consider only the most significantly enriched term, but a user can also select multiple. Our method also supports custom input SNPs. TAD_Pathways Software is available at https://github.com/greenelab/tad_pathways_pipeline.
Experimental knockdown of candidate genes
We investigated two candidate genes predicted by TAD_Pathways: ACP2 and DEAF1. We selected these genes because they are not the nearest gene and are not in the same LD block as the GWAS SNP (Supplementary Figures S1 and S2). Additionally, the genes were not previously known to impact human bone and thus represented potential novel research/treatment avenues. The corresponding BMD GWAS loci rs7932354 (11p11.2) and rs11602954 (11p15.5) were previously assigned to ARHGAP1 and BET1L, respectively.10
These genes were experimentally knocked down in a human fetal osteoblast (hFOB) cell line using a commercial siRNA reagent system in three temporally separated independent technical replicates. The influence of knockdown on gene expression (qPCR), cellular metabolism/proliferation (MTT), and early osteoblast differentiation (ALP) was evaluated within the first 4 days following siRNA transfection. All values are reported as mean±SD with statistical significance determined via two-way homoscedastic Student’s t-tests (*P≤0.05, #P≤0.10, NS='not significant'). Complete experimental methods are included in the Supplementary Information.
Results
TAD_Pathways reveals candidate genes within phenotype-associated TADs
We applied TAD_Pathways to BMD GWAS results derived from replication-requiring journals (see Supplementary Information publications). GWAS curation resulted in the aggregation of 70 unique BMD SNPs. TAD_Pathways implicated ‘Skeletal System Development’ as the top-ranked pathway (Benjamini–Hochberg adjusted P=1.02 × 10−5). For full BMD TAD_Pathways results refer to Supplementary Table S1. Many candidates were not the nearest gene to the GWAS signal and several had independent eQTL support (Supplementary Table S2).
We compared TAD boundary gene aggregation to nearest-gene and LD windows (r2>0.4). The aggregated gene lists included different gene sets, with TAD boundaries aggregating the most genes (Supplementary Figure S3A). We also applied a pathway analysis to each gene set, and the top pathway for all methods was ‘Skeletal System Development’. TAD_Pathways identified 38 total candidate genes and 17 unique genes not discovered by either nearest-gene or LD approaches (Supplementary Figure S3B).
siRNA knockdown of candidate genes in osteoblasts
We targeted the expression of four genes in vitro using siRNA and assessed transcriptional knockdown efficiency (Figure 2). We noted variation across the three controls, with the scrambled siRNA control altering expression of OCN (osteocalcin), IBSP (bone sialoprotein), TNAP and BET1L (P<0.05). Relative to the scrambled siRNA control, OCN was downregulated in all siRNA groups (P<0.05), except for BET1L siRNA (P=0.122). OSX, IBSP and TNAP were not significantly altered by any siRNA treatment (Figure 2).
Real-time PCR of osteoblast differentiation genes and GWAS/TAD hits in hFOB cells. siRNA was used to knockdown expression of TNAP (positive control), ARHGAP1, ACP2, BET1L and DEAF1. Relative expression of the osteoblast marker genes OSX, OCN and IBSP suggests that GWAS/TAD hits are not major regulators of bone differentiation in this model. Red bars highlight specificity of each siRNA knockdown. Values represent mean±SD. Statistical significance relative to the scrambled siRNA control is annotated as: *P≤0.05 and #P≤0.10 using a two-tailed Student’s t-test.
Metabolic and osteoblastic activity of TAD_Pathways gene predictions
Treatment with ACP2 siRNA led to a 66.0% reduction in MTT metabolic activity versus the scrambled siRNA control (P=0.012). ARHGAP1 siRNA caused a 38.8% reduction (P=0.088). siRNA targeted against TNAP, BET1L or DEAF1 did not alter MTT metabolic activity (Figure 3a).
Validating two TAD_Pathways predictions for BMD GWAS hits on hFOB cells. siRNA was used to knockdown expression of TNAP, ARHGAP1, ACP2, BET1L and DEAF1. (a) Knockdown of ACP2 decreases cellular metabolic activity, demonstrated using an MTT assay. (b) ALP staining and quantitation indicates that knockdown of TNAP or ACP2 inhibits performance in an osteoblast differentiation assay. Values represent mean±SD. Statistical significance relative to the scrambled siRNA control is annotated as: *P≤0.05 and #P≤0.10 using a two-tailed Student’s t-test.
ALP is highly expressed in osteoblasts: disruption of proliferation or osteoblast differentiation results in ALP downregulation. TNAP siRNA significantly reduced ALP intensity by 5.98±1.77 units versus the scrambled siRNA control (P=0.006). ACP2 siRNA also significantly reduced ALP intensity by 8.74±2.11 (P=0.003). The control stained less intensely than untreated or transfection reagent controls, but this did not reach statistical significance (0.05<P<0.10) (Figure 3b).
Discussion
We show that TAD_Pathways can reveal functional gene to intermediate phenotype relationships using BMD. Several of the TAD_Pathways genes, such as LRP5, are bona fide BMD genes already identified by several methods, thus providing positive controls. However, several BMD GWAS signals do not have obvious nearest-gene associations with bone. Our results suggest that a nearby gene, ACP2, and not the nearest gene, ARHGAP1, regulates osteoblast proliferation/viability. There is modest previous evidence that ACP2 impacts bone in mouse models11 and is thus a promising candidate for follow-up studies.
There are several limitations to our approach. Publication biases from pathway curation present challenges.12 To lessen this bias, we include computationally predicted GO annotations. We used TAD boundaries defined by Dixon et al.,13 whereas increased Hi-C resolution reduced TAD sizes. Despite our method using larger TADs, we still identify relevant pathways. However, the method will fail in diseases instigated by aberrant looping. We were also concerned that TAD_Pathways works only with BMD. We applied TAD_Pathways to T2D and identify several candidate genes that are also not the nearest gene (see Supplementary Table S3). Moreover, the experimental validation was performed in a tetraploid in vitro cell culture system, which may compensate for gene knockdown. While TAD_Pathways identified several candidate genes, we only examined two, and our validation approach does not directly interrogate each SNP.
One of the investigated GWAS SNPs, rs7932354, located in the ARHGAP1 promoter, is an eQTL for ARHGAP1 in several GTEx tissues14 and is associated with epigenetic marks and alternative genes in HaploReg.15 However, none of these tissues are bone related and our screen implicates ACP2 and not ARHGAP1 in osteoblast processes. Furthermore, LRP4 and PACSIN3 also fall within the rs7932354 TAD and LD block (Supplementary Figure S1). Both genes are associated with bone.16, 17 Therefore, TAD_Pathways revealed additional genes that would otherwise have been overlooked by alternative methods.
In conclusion, TAD_Pathways can be used as a candidate gene discovery tool through the leveraging of features of chromatin looping. TAD_Pathways is different from previous approaches, such as DEPICT18 and MAGENTA,19 because it only requires the trait as user input and can be performed rapidly. Our method builds solely from publicly available GWAS and TAD boundaries. TAD_Pathways overcomes SNP abundance-related gene selection biases pervasive in previous methods by aggregating SNPs directly to TADs instead of genes.20 We believe TAD_Pathways and algorithms that leverage 3D genomic structure will aid in the discovery of novel disease features. A Supplementary Video is available at the European Journal of Human Genetics website.
References
Welter D, MacArthur J, Morales J et al: The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res 2014; 42: D1001–D1006.
Brodie A, Azaria JR, Ofran Y : How far from the SNP may the causative genes be? Nucleic Acids Res 2016; 44: 6046–6054.
Claussnitzer M, Dankel SN, Kim K-H et al: FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med 2015; 373: 895–907.
Xia Q, Chesi A, Manduchi E et al: The type 2 diabetes presumed causal variant within TCF7L2 resides in an element that controls the expression of ACSL5. Diabetologia 2016; 59: 2360–2368.
Lieberman-Aiden E, van Berkum NL, Williams L et al: Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009; 326: 289–293.
Dixon JR, Selvaraj S, Yue F et al: Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012; 485: 376–380.
Ho JWK, Jung YL, Liu T et al: Comparative analysis of metazoan chromatin organization. Nature 2014; 512: 449–452.
Wang J, Duncan D, Shi Z, Zhang B : WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res 2013 41: W77–W83.
Ashburner M, Ball CA, Blake JA et al: Gene Ontology: tool for the unification of biology. Nat Genet 2000; 25: 25–29.
Estrada K, Styrkarsdottir U, Evangelou E et al: Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet 2012; 44: 491–501.
Suter A, Everts V, Boyde A et al: Overlapping functions of lysosomal acid phosphatase (LAP) and tartrate-resistant acid phosphatase (Acp5) revealed by doubly deficient mice. Dev Camb Engl 2001; 128: 4899–4910.
Greene CS, Troyanskaya OG : Accurate evaluation and analysis of functional genomics data and methods: Accurate evaluation and analysis of functional genomics data and methods. Ann N Y Acad Sci 2012; 1260: 95–100.
Rao SSP, Huntley MH, Durand NC et al: A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014; 159: 1665–1680.
Aguet F, Brown AA, Castel S et al Local Genetic Effects on Gene Expression Across 44 Human Tissues, 2016. Available at: http://biorxiv.org/lookup/doi/10.1101/074450 (last accessed 3 January 2017).
Ward LD, Kellis M : HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 2012; 40: D930–D934.
Boudin E, Steenackers E, de Freitas F et al: A common LRP4 haplotype is associated with bone mineral density and hip geometry in men – data from the Odense Androgen Study (OAS). Bone 2013; 53: 414–420.
Blake JA, Eppig JT, Kadin JA, Richardson JE, Smith CL, Bult CJ : Mouse Genome Database (MGD)-2017: community knowledge resource for the laboratory mouse. Nucleic Acids Res 2017; 45: D723–D729.
Pers TH, Karjalainen JM, Chan Y et al: Biological interpretation of genome-wide association studies using predicted gene functions. Nat Commun 2015; 6: 5890.
Segrè AV, Consortium D, Investigators M et al: Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet 2010; 6: e1001058.
Greene CS, Himmelstein DS : Genetic association-guided analysis of gene networks for the study of complex traits. Circ Cardiovasc Genet 2016; 9: 179–184.
Acknowledgements
Hannah E Sexton and Troy L Mitchell assisted in optimizing siRNA transfection conditions. Daniel Himmelstein and Amy Campbell performed analytical code review. This work was supported by the Genomics and Computational Biology Graduate program at the University of Pennsylvania (to GPW); the Gordon and Betty Moore Foundation’s Data Driven Discovery Initiative (grant number GBMF 4552 to CSG); the National Institute of Dental and Craniofacial Research (NIH grant number F32DE026346 to DWY). SFAG is supported by the Daniel B Burke Endowed Chair for Diabetes Research. All data used to construct the TAD_Pathways approach are publically available data sets. All the softwares used to develop this approach are publically available in a GitHub repository (http://github.com/greenelab/tad_pathways_pipeline). We also provide a docker image (https://hub.docker.com/r/gregway/tad_pathways/) and archive the GitHub Software on Zenodo (https://zenodo.org/record/254190).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Way, G., Youngstrom, D., Hankenson, K. et al. Implicating candidate genes at GWAS signals by leveraging topologically associating domains. Eur J Hum Genet 25, 1286–1289 (2017). https://doi.org/10.1038/ejhg.2017.108
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2017.108
This article is cited by
-
Genome-wide association study implicates novel loci and reveals candidate effector genes for longitudinal pediatric bone accrual
Genome Biology (2021)
-
Effect of 6p21 region on lung function is modified by smoking: a genome-wide interaction study
Scientific Reports (2020)
-
Chromothripsis, a credible chromosomal mechanism in evolutionary process
Chromosoma (2019)
-
Maternal and fetal genetic effects on birth weight and their relevance to cardio-metabolic risk factors
Nature Genetics (2019)
-
Leveraging epigenomics and contactomics data to investigate SNP pairs in GWAS
Human Genetics (2018)