Abstract
Linkage disequilibrium (LD) is the non-random distribution of alleles across the genome, and it can create serious problems for modern linkage studies. In particular, computational feasibility is often obtained at the expense of power, precision, and/or accuracy. In our new approach, we combine linkage results over multiple marker subsets to provide fast, efficient, and robust analyses, without compromising power, precision, or accuracy. Allele frequencies and LD in the densely spaced markers are used to construct subsamples that are highly informative for linkage. We have tested our approach extensively, and implemented it in the software package EAGLET (Efficient Analysis of Genetic Linkage: Estimation and Testing). Relative to several commonly used methods we show that EAGLET has increased power to detect disease genes across a range of trait models, LD patterns, and family structures using both simulated and real data. In particular, when the underlying LD pattern is derived from real data, we find that EAGLET outperforms several commonly used linkage methods. In-depth analysis of family data, simulated with linkage and under the real-data derived LD pattern, showed that EAGLET had 78.1% power to detect a dominant disease with incomplete penetrance, whereas the method that uses one marker per cM had 69.7% power, and the cluster-based approach implemented in MERLIN had 76.7% power. In this same setting, EAGLET was three times faster than MERLIN, and it narrowed the MERLIN-based confidence interval for trait location by 29%. Overall, EAGLET gives researchers a fast, accurate, and powerful new tool for analyzing high-throughput linkage data, and large extended families are easily accommodated.
Similar content being viewed by others
INTRODUCTION
Although genome-wide association studies have identified more than 400 genomic regions associated with risk for complex disease; these associations only account for a small fraction of the overall genetic component of risk.1, 2 In contrast, the successes of linkage analysis3, 4, 5, 6 have led to the identification of genetic factors with large effects. As linkage studies began to genotype individuals at thousands of common single nucleotide polymorphisms (SNPs), a handful of genetic mutations with modest effects were also identified.7, 8 At present, most genetic studies are genotyping individuals at millions of sites, often comprising a mixture of both common and rare variants, but current multipoint linkage methods cannot analyze such high-throughput data efficiently. Therefore, the power to detect pathogenic mutations may be substantially increased by making more efficient use of this valuable resource.
High-throughput linkage data exhibit linkage disequilibrium (LD), a phenomenon whereby the alleles at neighboring loci are correlated. When analyzing such data, multipoint methods usually sacrifice efficiency and/or accuracy for the sake of computational feasibility. For example, ignoring LD is perhaps the simplest means of achieving computational feasibility, but this can inflate type I error rates.9, 10, 11, 12 Furthermore, ignoring LD tends to distort estimates of trait locus position when linkage is present.13 As an alternative to ignoring LD, most multipoint methods either significantly reduce LD by removing markers14, 15, 16 or model LD directly within the analysis.17, 18, 19, 20 For methods that remove markers, such as single subsample-based methods, there are two main disadvantages. First, the analysis is often inefficient because a large number (usually the vast majority) of the high-throughput genotypes are typically ignored. Second, there is little assurance that the chosen subsample is optimal. There are also drawbacks for methods that model LD. Typically, the assumptions that underlie these methods (such as independently segregating haplotype blocks, or a first-order Markov process for allelic states along the chromosome) are not justified; and, the implicit misfit of the model often leads to a loss in power. Moreover, some of these methods can be computationally impractical, even for simple designs such as affected sibpairs (ASPs).
We present a new test for linkage that uses all of the available genotype data by combining linkage results over multiple subsamples. The allele frequencies and LD of the dense genotype data are used to construct subsamples that are highly informative for linkage. We have implemented our new approach in the software package: EAGLET (Efficient Analysis of Genetic Linkage: Estimation and Testing), which is freely available from the Web (see Web Resources for the URL). Note that once linkage has been established, EAGLET can construct narrow and accurate confidence intervals for the location of the unknown disease gene.13 Moreover, all of EAGLET's many features (eg, linkage detection, trait location estimation, pairwise LD estimation, and family-based meta-analysis) can be applied to large extended families.
MATERIALS AND METHODS
Our method uses information from multiple subsamples of dense SNP linkage data to detect disease genes in the presence of LD. Each subsample uses a different set of SNPs, but retains all family members. As the markers within each subsample are chosen on the basis of their relative heterozygosity, the subsamples are highly informative for linkage. Furthermore, to account for LD, we place an upper bound on the pairwise LD (as measured by r2 or D′) between adjacent markers in a subsample. When LD is present in the high-throughput data, the power to detect linkage depends crucially on the degree to which the linkage equilibrium (LE) assumption is violated within each subsample. We define subsample size as the number of markers in a subsample, and show that power is a function of the expected subsample size divided by the total number of markers on the chromosome; we denote this quantity by λ. To understand why power depends on λ, consider the case when λ≈1. In this event, the markers within each subsample tend to be closely spaced and in high LD, which grossly violates the LE assumption and reduces power. Similarly, when λ≈0, the markers tend to be sparsely spaced within each subsample, which can significantly reduce the evidence for linkage, and hence the power. In general, power is maximized for λ∈(0, 1).
Subsampling algorithm
Let the pair (T, G) denote all trait and high-throughput genotype data observed on some (possibly all) members of one or more families. For a given chromosome, define G≡(G1,…, GN) as the collection of single-locus genotypes, where Gi denotes all genotypes observed at the ith marker for i=1, 2, …, N. Furthermore, let 𝒢 denote a subsample of the high-throughput genotype data G. We use the following algorithm (described below) to generate a random subsample. For any chosen value of λ, the goal of the algorithm is to select a subsample that is highly informative for linkage.
Step 1: Choose a random marker from the N available markers. Make this marker the most recently included marker, and add it to the subsample.
Step 2: Find the first available marker downstream of the most recently included marker such that the LD between the pair is less than the marker-specific threshold, κ. Make this marker the proposed marker. Note that the marker-specific threshold κ, which bounds the degree to which the LE assumption is violated, is determined implicitly by λ. A more precise definition of κ is given below.
Step 3: Incorporate the proposed marker into the subsample with probability μ, where μ depends on κ, the heterozygosity of the proposed marker, and the LD between the proposed marker and the most recently included marker. For a detailed discussion of how EAGLET computes μ see Appendix A.
Step 4: If the proposed marker is incorporated, it becomes the most recently included marker. Otherwise, it becomes unavailable.
Step 5: Return to Step 2 until every marker downstream of the most recently included marker is either unavailable or has LD with the most recently included marker that is greater than or equal to κ.
An analogous procedure starting at Step 2 is used to select markers upstream of the very first marker chosen in Step 1. This completes construction of a random subsample.
In our implementation, the marker-specific value κ=κo when the marker in question has heterozygosity equal to the mean heterozygosity of the sample. Note that the user-defined constant κo is intended to represent an upper bound on the average pairwise LD, where the average is taken across adjacent intervals of a subsample. In contrast, the marker-specific value κ varies as the heterozygosity of the marker differs from the mean heterozygosity of the sample. In particular, more informative markers are proposed more often than less informative markers. For a detailed discussion of the computation of κ, see Appendix A. Furthermore, since the acceptance probability μ decreases as the number of markers with similar heterozygosity increases, proposed markers that are common are less likely to be incorporated than proposed markers that are rare. By varying the acceptance probabilities in this way, the resultant subsample retains most of the information about linkage while maintaining a relatively small amount of LD. For a detailed discussion of the computation of acceptance probabilities, see Appendix A.
Test statistic and power
In addition to the use of multiple subsamples, EAGLET also relies on the fast and efficient computation of the Kong and Cox LOD21 Zlr2(x), a so-called ‘model free’ LOD score22 evaluated at location x along the chromosome. Our test statistic is the average, over subsamples, of the max Zlr2 statistic. In what follows, we suppress the symbolic representation of trait data T, as all distributions P (·) and likelihoods L(·) are conditional on T.
As the biological relationship between a pair of individuals determines the expected number of alleles that they share identical by descent for any location x in the genome, sharing among a set of affected relatives in excess of this expectation is usually interpreted as evidence for linkage. A commonly used test statistic that quantifies the amount of excess sharing is max Zlr2, defined as

where δ̂(x) measures the departure from expected sharing at location x, and ρ is a known vector of nuisance parameters containing recombination rates and allele frequencies specific to 𝒢. To make use of all of the high-throughput genotype data, our test statistic

is approximated by the average of max Zlr2 over K random subsamples.
Recall that for computational reasons, LD is almost always ignored when computing max Zlr2. This means that in unlinked regions of strong LD, the apparent sharing among relatives is inflated, especially if the genotypes of founders are missing. If, in addition to missing data on founders, the study is also enriched for affected members (eg, ASPs), then the increase in apparent sharing will be misinterpreted as evidence for linkage. However, by retaining small amounts of LD, it is possible to increase power without increasing the type I error. For example, when there is linkage, the power to detect a disease gene from a subsample in LE is generally increased by adding markers, even though this introduces LD that is subsequently ignored in the analysis. This happens because the negative effect of model misspecification (ie, assuming an LE model when LD is present) is often outweighed by the additional information obtained for linkage. Of course, if the subsample retains every marker of the dense genotype data G, then the LD is high, the model misspecification is severe, and power is typically low. Consequently, the power of a subsample is generally maximized for more than two but less than N markers. Moreover, for the methods and simulations considered in this paper, power is always maximized for λ∈[2/N, 1], where λ is the expected proportion of markers along the chromosome per subsample.
Data description
To assess the power of our proposed method, and to clarify the relative importance of several factors that influence the analysis of high-throughput linkage studies, we estimated the power of five different methods across nine different scenarios. The five methods, which we describe below, are implemented in the programs: EAGLET, MLOD, SLOD, EDIST, and MERLIN. The method used in MLOD is similar to EAGLET in that it also combines information across multiple subsamples, but with MLOD, the markers within each subsample are chosen on the basis of distance (genetic or physical). SLOD is also similar to EAGLET as it uses LD to choose markers, but SLOD only uses a single subsample. EDIST (which is a special case of MLOD) uses a single subsample, but markers are chosen to be equidistant (≈ one marker per cM). Finally, MERLIN assumes that SNPs in high LD form independent blocks, and that the alleles of each block are non-recombining haplotypes. The nine different simulation scenarios (which we describe below) encompass different patterns of LD, modes of inheritance, genetic effects, and family structures. The dense SNP linkage data were simulated with LD using the program CALEB (see Web References for the URL), and power (as a function of λ) was estimated from 600 replicates for the first eight scenarios and 1200 replicates for the ninth scenario. To ensure that all methods have approximately 5% type I error, we used 3000 realizations under the null hypothesis of no linkage to estimate the correct critical value for each combination of LD pattern and sample size.
We generated dense SNP linkage data for the nine different scenarios by changing the following factors: LD pattern, genetic effect, mode of inheritance (MOI), and family structure. We considered an LD pattern that consisted of three equi-length blocks containing ≈ 66 SNPs per block with alternating LD across blocks (as measured through D′). Within each block the pairwise D′ was 0.9, 0.1, and 0.9, respectively. This constitutes an extreme pattern of strong-weak-strong LD. We also considered an empirical LD pattern that was estimated from the dense SNP data of a real, genome-wide linkage scan.23 We used two levels of genetic effect with sibling relative risks of 1.5 and 2.0, and two modes of inheritance: dominant (DOM) and recessive (REC). For each combination, the family structures were ASPs with missing data on the parents, and a trait locus was positioned in the middle of 198 SNPs spread evenly along a 99-cM map. For the last scenario, we simulated the strong, dominant trait model using the empirical LD pattern on affected sibling trios (ASTs) to examine the effect of adding an affected sibling. In every scenario, the trait locus was positioned in between two regions of relatively high LD.
RESULTS
We used 4800 replicates of dense SNP ASP data to estimate power for five different multipoint linkage methods across eight different scenarios. In Tables 1 and 2, we report the maximum power, which was maximized over λ, for each of the eight scenarios. For the AST (trio) data, we simulated 500 replicates and estimated the power of all five methods for a single scenario (Table 3). For completeness, we also computed the 95% confidence intervals for trait location for EAGLET, MERLIN, and EDIST.
Table 1 shows that when data are simulated with the empirical LD pattern (which was estimated from an actual genome scan), EAGLET slightly outperforms the other methods, and does so irrespective of the MOI, or the strength of the genetic effect. Similarly, EAGLET continues to maintain a slight advantage over the other methods, irrespective of whether data are simulated on ASPs or ASTs (Table 3). From Table 2, we see that when the trait locus is positioned between long stretches of LD (D′=0.9), as in the extreme LD pattern with alternating blocks of LD: 0.9–0.1–0.9, EAGLET is comparable to MERLIN, but that the two distance-based methods MLOD and EDIST have noticeably less power. For example, when the sibling relative risk is 2.0, and the MOI is DOM, the power of EDIST and MLOD is only 13.8% and 19.7%, respectively; whereas, the power of EAGLET and MERLIN is 27.7% and 28.5%, respectively. Collectively, these results show that EAGLET has high power and is robust over the range of scenarios explored. In addition, EAGLET was generally faster than MERLIN, and for small values of λ, analyses with MERLIN were often computationally infeasible.
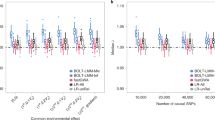
In Figure 1b, data from 1200 replicates with 200 ASTs per replicate were used to estimate the power functions of EAGLET, MERLIN, MLOD, and SLOD. Recall that power depends on λ the expected proportion of SNPs per subsample. Figure 1b shows that power is maximized in the interior for all methods. In addition, EAGLET's 95% confidence interval for trait location: (10.37, 55.96) is much tighter than the 95% confidence interval based on MERLIN: (11.06, 75.63), resulting in a 29% reduction in length (P=0.02). EAGLET also computes the average LOD score curve (ALOD), which is shown alongside the LOD score curves of MERLIN and SLOD in Figure 1a. The maximum ALOD has nearly identical power to EAGLET (data not shown), and it can be used to summarize the evidence for linkage along the chromosome.
(a) Shows the Kong and Cox LOD score curves and the 95% confidence intervals for trait locus position for three methods: EAGLET (solid black), MERLIN (solid gray), and EDIST (dotted black). (b) Shows power as a function of the expected proportion of SNPs for EAGLET (solid black), MERLIN (solid gray), MLOD (dotted gray), and SLOD (dashed gray). The dotted black line represents the Kong and Cox LOD for EDIST computed with roughly one SNP per cM.
DISCUSSION
EAGLET provides fast, efficient, and accurate analyses of dense SNP linkage data. What is equally important is that once linkage is established, EAGLET also yields tight confidence intervals of trait location with asymptotically valid coverage, thereby reducing the length of the segment that must be explored to identify the gene. EAGLET computes the ALOD, which provides a valuable summary of the evidence for linkage at position x that accounts for the variation across subsamples, and protects against the negative influence of data artifacts (eg, undetected map and genotype errors). Furthermore, as EAGLET does not make any assumptions about the underlying correlation structure, it is robust to the different patterns of LD in the genome.
In addition to the methods and LD patterns addressed here, we also compared the power of EAGLET with six other subsample-based methods (including some that used a weighted average of the information in each subsample) under three alternative LD patterns. EAGLET yielded robust performance across all scenarios, achieving power as high as, or better than, any of the alternative methods (data not shown). Note that of the methods discussed in this paper, MLOD is almost identical to the method proposed by Bacanu,14 and that SLOD is very similar to the method proposed by Bellenguez.16 Moreover, although all of these methods were designed for high-throughput SNP data, they are in principle, applicable to data sets containing single nucleotide variants as well.
For large samples, it is clear that accounting for variation across subsamples, and choosing subsamples on the basis of LD, yields tighter confidence intervals for trait locus position when compared with existing methods.13 However, it is only here (within the context of linkage detection) that we have explored the relative contribution of each factor separately. For example, it is usually better to choose a subsample on the basis of LD (eg, SLOD), than it is to combine information across subsamples chosen on the basis of distance (eg, MLOD). As EAGLET does both (ie, it uses LD to choose its subsample and it combines information across subsamples), it outperforms both SLOD and MLOD.
With the emerging promise of affordable whole-exome and/or whole-genome sequencing, many researchers hope to identify pathogenic mutations by examining the co-inheritance of rare variants with disease.24 However, the vast majority of rare variants (>99%) do not occur in exons, and the typical number of rare variants per genome (≈ 3.5 million) is staggering in comparison with the number of samples that project budgets can afford to resequence.25 Therefore, any practical implementation of a whole-genome sequencing approach (now and in the foreseeable future) will benefit substantially from complementary methods that allow researchers to prioritize rare variants (eg, candidate genes and predicted gene pathways). To the extent that multipoint linkage analysis identifies linked and unlinked regions of the genome, it provides an objective means of prioritizing rare variants and is potentially an extremely useful complement to whole-genome sequencing.
Until now, most multipoint linkage methods that account for LD have focused on minimizing LD. However, we have demonstrated empirically that power is maximized when the expected proportion of markers per subsample (denoted λ) is between zero and one. Therefore, when analyzing dense high-throughput linkage data, LD should not be completely removed nor should every marker be retained. This is why we maximized power over λ for each of the methods considered. However, when analyzing real data, this maximization may be computationally demanding or altogether infeasible for certain study designs. This means that for many researchers, the power of MERLIN, SLOD, and MLOD, is likely to be lower than what we report here, because many researchers will not know the optimal value of λ a priori. Currently, we are in the process of extending EAGLET to estimate the optimal value of λ and to incorporate parametric linkage statistics, so that researchers can begin to realize the full potential of high-throughput linkage data.
References
Goldstein DB : Common genetic variation and human traits. N Engl J Med 2009; 360: 1696–1698.
Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein D : Rare variants create synthetic genome-wide associations. PLoS Biol 2010; 8: e1000294.
Zengerling S, Tsui LC, Grzeschik KH, Olek K, Riordan JR, Buchwald M : Mapping of DNA markers linked to the cystic fibrosis locus on the long arm of chromosome 7. Am J Hum Genet 1987; 40: 228–236.
Van Broeckhoven C, Backhovens H, Cruts M et al: Mapping of a gene predisposing to early-onset Alzheimer's disease to chromosome 14q24.3. Nat Genet 1992; 2: 335–339.
Ottman R, Risch N, Hauser WA et al: Localization of a gene for partial epilepsy to chromosome 10q. Nat Genet 1995; 10: 56–60.
Hampe J, Frenzel H, Mirza MM et al: Evidence for a NOD2-independent susceptibility locus for inflammatory bowel disease on chromosome 16p. Proc Natl Acad Sci USA 2002; 99: 321–326.
Reynisdottir I, Thorleifsson G, Benediktsson R et al: Localization of a susceptibility gene for type 2 diabetes to chromosome 5q34-q35. Am J Hum Genet 2003; 73: 323–335.
Zhou K, Dempfle A, Arcos-Burgos M et al: Meta-analysis of genome-wide linkage scans of attention deficit hyperactivity disorder. Am J Med Genet B (Neuropsychiatr Genet) 2008; 147B: 1392–1398.
Huang Q, Shete S, Amos CI : Ignoring linkage disequilibrium among tightly linked markers induces false-positive evidence of linkage for affected sib pair analysis. Am J Hum Genet 2004; 75: 1106–1112.
Boyles AL, Scott WK, Martin ER et al: Linkage disequilibrium inflates type I error rates in multipoint linkage analysis when parental genotypes are missing. Hum Hered 2005; 59: 220–227.
Goode EL, Badzioch MD, Jarvik GP : Bias of allele-sharing linkage statistics in the presence of intermarker linkage disequilibrium. BMC Genet 2005; 6: S82.
Kim Y, Duggal P, Gillanders EM, Kim H, Bailey-Wilson JE : Examining the effect of linkage disequilibrium between markers on the type I error rate and power of nonparametric multipoint linkage analysis of two-generation and multigenerational pedigrees in the presence of missing genotype data. Genet Epidemiol 2008; 32: 41–51.
Stewart WCL, Peljto AL, Greenberg DA : Multiple subsampling of dense SNP data localizes disease genes with increased precision. Hum Hered 2010; 69: 152–159.
Bacanu S : Multipoint linkage analysis for a very dense set of markers. Bioinformatics 2005; 29: 195–203.
Webb EL, Sellick GS, Houlston RS : SNPLINK: multipoint linkage analysis of densely distributed SNP data incorporating automated linkage disequilibrium removal. Bioinformatics 2005; 21: 3060–3061.
Bellenguez C, Ober C, Bourgain C : Linkage analysis with dense SNP maps in isolated populations. Hum Hered 2009; 68: 87–97.
Abecasis GR, Wigginton JE : Handling marker-marker linkage disequilibrium: pedigree analysis with clustered markers. Am J Hum Genet 2005; 77: 754–767.
Allen-Brady K, Horne BD, Malhotra A, Teerlink C, Camp NJ, Thomas A : Analysis of high-density single-nucleotide polymorphism data: three novel methods that control for linkage disequilibrium between markers in a linkage analysis. BMC Proc 2007; 1: S160.
Albers C, Kappen HJ : Modeling linkage disequilibrium in exact linkage computations: a comparison of first-order markov approaches and the clustered-markers approach. BMC Proc 2007; 1: S159.
Kurbasic A, Hossjer O : A general method for linkage disequilibrium correction for multipoint linkage and association. Genet Epidemiol 2008; 32: 647–657.
Kong A, Cox NJ : Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 1997; 61: 1179–1188.
Whittemore AS : Genome scanning for linkage: an overview. Am J Hum Genet 1996; 59: 704–716.
Rodriguez-Murillo L, Subaran R, Stewart WCL et al: Novel loci interacting epistatically with bone morphogenetic protein receptor 2 cause familial pulmonary arterial hypertension. J Heart Lung Transplant 2009; 29: 174–180.
Cirulli ET, Goldstein DB : Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet 2010; 11: 415–425.
Pelak K, Shianna KV, Ge D et al: The characterization of twenty sequenced human genomes. PloS Genet 2010; 6: e1001111.
Acknowledgements
We would like to acknowledge the support of the Calderone City Health Award, and of the National Institutes of Health: NLO60056, MH48858, MH65213, DKO31175, DKO31813, DA026983, and NS070323.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Appendix A
Appendix A
THE COMPUTATION OF MARKER-SPECIFIC THRESHOLDS AND ACCEPTANCE PROBABILITIES
To facilitate the exposition of marker-specific thresholds and acceptance probabilities, we encourage readers to consider the following shopping problem. Imagine that you have a fixed amount of money and that you are to purchase an unspecified number of items from a collection of items arranged in a line. Upon inspecting each item, you must decide to purchase or not to purchase. Ultimately, your goal is to end up with the best subcollection of items. Now, this simple shopping problem is extremely similar to the problem of constructing a highly informative subsample from a modern linkage data set. In particular, the items that are arranged in a line can be viewed as the dense SNPs, and the items that are eventually purchased are analogous to a highly informative subsample. However, instead of spending money to obtain items, SNPs are purchased (ie, incorporated into the subsample) by admitting different amounts of LD. Similarly, just as each item has a value, each SNP has a value. Moreover, the entire subcollection of items has a value, as does the subsample. In particular, each subsample provides power to detect linkage, and this is what we want to maximize. Finally, as is generally the case with shopping, a deal (ie, purchasing an item for less than it is actually worth) is usually advantageous. Let hi be the heterozygosity of SNPi for i=1,…, N, where N is the number of SNPs in the original high-throughput genotype data. Let h̄ be the average heterozygosity across all SNPs, and let κo denote the average pairwise LD across adjacent intervals of a subsample for a typical modern linkage study (eg, r2=0.01). Now, on the basis of hi, assign each SNP to one of 10 mutually exclusive categories: (0, 0.05), (0.05, 0.1), …, (0.40, 0.45), (0.45, 0.5). Further, let pi be the proportion of SNPs belonging to the ith category. To decide how much LD we are willing to admit, we compute a marker-specific threshold κi that is less than κo if hi<h̄ and greater than κo if hi>h̄. Basically, we are willing to pay more for SNPs that are, on average, more informative. Therefore, given a symmetric window (a, b) centered around κo with 0<a, b<1, κi is interpolated from the ratio of (hi−h̄) to (0.5−h̄). This completes the computation of marker-specific thresholds. Now, let Δ denote the LD between the most recently included SNP and the proposed SNP (see Subsampling Algorithm 2.1). Let j∈{1,…, 10} denote the category in which the proposed SNP falls. If pj>0.1 then the proposed SNP is accepted with probability

If pj≤0.1 then the proposed SNP is accepted with probability

Note that, (κi−Δ)/κi measures the strength of the deal, and given that a SNP has been proposed, common SNPs (ie, SNPs with pj>0.1) are accepted less often than rare SNPs. This completes the computation of acceptance probabilities.
Rights and permissions
About this article
Cite this article
Stewart, W., Drill, E. & Greenberg, D. Finding disease genes: a fast and flexible approach for analyzing high-throughput data. Eur J Hum Genet 19, 1090–1094 (2011). https://doi.org/10.1038/ejhg.2011.81
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2011.81