Genetic differences between individuals can affect how they respond to drugs.Credit: Jekesai Njikizana/AFP/Getty
How a person will respond to a drug is, in part, determined by their genetics. Africa holds the world’s most genetically diverse human population, and the United Nations estimates that, by 2050, the continent will be home to nearly 25% of the world’s people. Yet pharmacogenomics research — studies of how genetic variation plays into drug responses — is sorely lacking in African populations.
Less than 5% of the data in the pharmaco-genomics database PharmGKB are from African populations1. And of more than 300 drugs for which the US Food and Drug Administration provides pharmacogenetic advice, only 15 have been studied in African groups2.
Artificial intelligence (AI) can help to close the gap. AI models trained to identify pharmaco-genetic variants — DNA mutations that might affect how a drug acts — are emerging in many countries in the global north. But a dearth of genetic data for African populations, along with a lack of training and infrastructure, is holding up the use of such models in Africa. Here, we outline ways to overcome these hurdles.
Africa needs pharmacogenetics
Pharmacogenetic data have two key purposes. First, they can be used to select the best drugs for an individual person — for example, people with a pharmacogenetic mutation in an immune-response gene called HLA-B are hypersensitive to the antiviral drug abacavir, and should therefore be prescribed alternatives3. Second, such information can be used to refine the dose of existing drugs. For instance, a mutation in the gene CYP2C9, which encodes a cytochrome P450 enzyme involved in drug metabolism, results in reduced breakdown of the commonly used blood thinner warfarin. People who have this variant should be given a lower dose of the drug to prevent a build-up of unmetabolized warfarin in the body that would increase the risk of a haemorrhage4.
AI can help to speed up drug discovery — but only if we give it the right data
But pharmacogenetic information generated in the global north is not always relevant to African populations, because genetic variants are found at varying frequencies in different ethnogeographical groups. Research into variants that specifically affect drug responses in Africans living in Africa, and the African diaspora, is essential for several reasons.
First, a reliance on clinical data from the global north can put the health of Africans at risk. Take efavirenz. This promising HIV/AIDS drug was used successfully in the United States and Europe before being launched as a first-line treatment in Zimbabwe in 2015. But the dosing recommendations for Zimbabweans did not take into account that Africans are more likely than Europeans and Americans to carry a mutation in the gene CYP2B6. This mutation is associated with a range of side effects5. Whereas dizziness, irritability or headache were commonly reported side effects in European and US patients6, many people in Zimbabwe experienced hallucinations, anxiety and suicidal ideation when taking efavirenz7.

A researcher analyses results at a tuberculosis laboratory in Cotonou, Benin.Credit: Yanick Folly/AFP/Getty
Second, the effects of pharmacogenetic variants need to be considered alongside several other factors that play out differently in Africa compared with other world regions. For instance, each year, the 2.5 million people who contract tuberculosis in Africa are typically given the antimicrobial drug rifampicin, among other treatments. But rifampicin speeds up the body’s ability to metabolize drugs8, so if a person is taking medications for other conditions, their dosages will probably need to be modified. Lifestyle factors such as diet, which varies between populations, also affect drug metabolism, by altering the community of microorganisms in a person’s gut, which in turn affects how the body processes a drug. Research conducted outside Africa will probably fail to factor in these complexities.
Third, there are commercial incentives. The African population, coupled with the diaspora, represents a huge market share for drugs. Dosing and prescription adjustments to make drugs safe for these populations is likely to increase uptake, and so boost profits for global pharmaceutical companies.
AI on the horizon for Africa
Pharmacogenetic variants are hard to find — it can take vast swathes of clinical and genetic data to pinpoint a variant that is associated with a change in drug response. AI models are moving the field forwards by scouring the scientific literature for drug–gene connections that humans have missed.
Could Africa be the future for genomics research?
To identify more variants, the next step — building on large language models such as GPT-4 and LLaMA — is to train ‘foundation’ AI models that bring together several types of data for analysis. For pharmacogenetics, this information will include large-scale genetics resources such as biobanks; electronic health records containing medical text and treatment responses; clinical-trial reports and drug labels that capture adverse drug reactions and prescription recommendations; and in vivo and in vitro data about genetics and drug activity from the existing scientific literature.
Foundation models are already being developed for clinical science — for example, to identify biomarkers of cancer in imaging data9. We expect that an open-access foundation model with applications in pharmacogenetics will be available in the next year or two.
These foundation models will be biased towards countries in the global north, because their training data will come mainly from people of European descent (see ‘Data bias’). But researchers in Africa can take advantage of an approach called transfer learning, in which a trained foundation model is fine-tuned using a smaller data set — in this case, information specific to African populations. Transfer learning has been used successfully for image recognition, with a handful of labelled photos allowing the model to learn new patterns10. We are confident that there are already enough African data available for transfer learning to begin to identify pharmacogenetic variants.
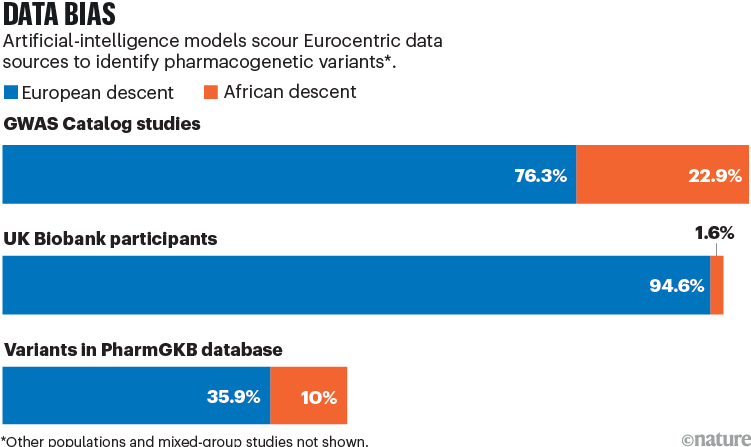
Sources: GWAS Catalog: https://go.nature.com/4CPZMJQ; UK Biobank: A. Fry et al. Am. J. Epidemiol. 186, 1026–1034 (2017); PharmGKB: https://go.nature.com/4A75YAX
Four steps to progress
African countries mostly rank low on the AI readiness index published by the UK consultancy Oxford Insights, with sub-Saharan Africa the worst-scoring world region when it comes to how ready governments are to implement AI in public services11. The following changes are needed to ensure that the scientific community in Africa is ready to harness transfer learning — and future AI tools for pharmacogenetic research (see also ‘The future of AI in pharmacogenetics’).
Train African researchers. Scientists in Africa are best positioned both to leverage knowledge of traditional medicine in Africa and to understand disease epidemiology in their regions. Africans, therefore, can best determine the research and data needed for effective drug discovery and drug tailoring on the continent.
International funders, research institutions and pharmaceutical companies must invest in training African researchers in AI and pharmacogenetics. Partnering with African initiatives such as Pharmacometrics Africa — a non-profit organization that provides training in clinical pharmacology — can help institutes to build local capacity.
Keep collecting data. Although transfer learning will be helpful for identifying pharmacogenetic variants that are common across Africa, many more genome sequences and focused clinical-trial results are needed to fully capture pharmacogenetic differences between African populations. Africa is not homogeneous — different ethnogeographical groups should be considered separately, in much the same way as biomedical research considers different European populations. The number of clinical trials on the continent is rising; future trials must include diverse cohorts of people, spanning multiple ethnicities.
Researchers wishing to conduct clinical trials across Africa currently need to apply to the health authorities of each country, each of which has different legal standards, fees and response timelines. When the African Medicines Agency becomes fully operational, clinical-trial regulation should become harmonized across the continent. With the date of this unknown, until then researchers can get help from the Clinical Trials Community — a platform hosted by the clinical software company Nuvoteq in Pretoria, South Africa, that provides up-to-date resources on the various regulatory and ethics requirements for conducting clinical trials in each country in Africa.
Invest in infrastructure and equipment. Genomics facilities are scarce in many African countries. A group of internationally renowned genomics researchers hopes to establish eight centres of excellence in genomics in Africa, each coordinated with a local academic unit and public-health facility. This initiative could bring world-class genomics research to Africa, generating the data needed to identify pharmacogenetic variants. To make this a reality, strong coordination between international funders, local governments and the Africa Centres for Disease Control and Prevention in Addis Ababa, Ethiopia, is needed to ensure that funding for the project materializes.
Allow patents on AI-generated inventions — for the good of science
And genomics tools such as pharmacogenetics testing kits — which analyse biological samples for thousands of mutations in genes involved in drug responses — must be tailored to Africans. This technology could help clinicians to adapt drug prescriptions to each patient’s genetic characteristics, bringing personalized medicine to Africa. Companies that make these tools, such as the US biotechnology firm Illumina, should work with African scientists to drive the inclusion of Africa-relevant variants as they are discovered.
Develop frameworks for data sharing and ethical research. It is unlikely that a single African institution, city or even country will have all the necessary components — human capacity, research infrastructure and data-collection sites — to be able to conduct world-class pharmacogenetics research. Data sharing is therefore needed.
But local researchers are concerned that openly sharing their data will put them at a disadvantage compared with colleagues in the global north, who might have more resources, infrastructure and skilled personnel to analyse the data and publish their findings. This reluctance is exacerbated by the fact that obtaining ethical approval to share and reuse samples is time-consuming — explaining and obtaining informed consent for genomics research often involves translating consent forms into local languages, for instance.
African scientists must build trusted research networks for sharing of data. These efforts will benefit from having continent-level legal and ethics frameworks for data sharing, enabling cross-country collaborations.
A good example of how data sharing across the continent can stimulate world-class research comes from the Pan-African Bioinformatics Network (H3ABioNet). This research consortium manages data storage and network infrastructure for the Human Heredity and Health in Africa (H3Africa) initiative — the largest effort to coordinate genomics research in Africa so far. The network has facilities in several African countries and well-defined data-submission and access policies for human genomics. It combines locally led management committees with training, good computational infrastructure and clear policies about data quality, allowing results to be deposited in a shared databank and incorporated into international databases such as the European Genome–phenome Archive.
When sharing of open data is restricted — in the case of a patient’s biomedical data, for instance — ‘decentralized’ AI algorithms can help. These are trained on data from numerous institutes, but in a way that allows each institute to keep its own data private. Such cooperative algorithms have been piloted in reference centres and hospitals in Europe and the United States for medical-imaging data.
The first steps towards realizing pharmacogenetics research in Africa are being taken, and AI can play a pivotal part in moving the field forwards. With successful capacity building, people in Africa will benefit from safer and more effective treatments, reducing the cost of health care on the continent. Without it, the disease-eradication goals outlined by the World Health Organization for the current decade are unlikely to be met.

 AI can help to speed up drug discovery — but only if we give it the right data
AI can help to speed up drug discovery — but only if we give it the right data
 Allow patents on AI-generated inventions — for the good of science
Allow patents on AI-generated inventions — for the good of science
 Neuron-D: Accelerating drug development with 3D neural models
Neuron-D: Accelerating drug development with 3D neural models
 Could Africa be the future for genomics research?
Could Africa be the future for genomics research?