- NEWS AND VIEWS
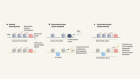
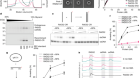
DNA-binding proteins meet their mismatch
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout
Nature 587, 199-200 (2020)
doi: https://doi.org/10.1038/d41586-020-02658-x
References
Afek, A. et al. Nature 587, 291–296 (2020).
Rohs, R. et al. Annu. Rev. Biochem. 79, 233–269 (2010).
Samee, M. A. H., Bruneau, B. G. & Pollard, K. S. Cell Syst. 8, 27–42 (2019).
Dien, V. T. et al. J. Am. Chem. Soc. 140, 16115–16123 (2018).
Zhang, W. H., Otting, G. & Jackson, C. J. Curr. Opin. Struct. Biol. 23, 581–587 (2013).
Lambert, S. A. et al. Cell 172, 650–665 (2018).

 Read the paper: DNA mismatches reveal conformational penalties in protein–DNA recognition
Read the paper: DNA mismatches reveal conformational penalties in protein–DNA recognition
 Smoking gun for a rare mutation mechanism
Smoking gun for a rare mutation mechanism
 Strands of evidence about cancer evolution
Strands of evidence about cancer evolution