- NEWS AND VIEWS
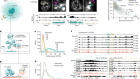
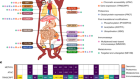
Extinct proteins resurrected to reconstruct the evolution of vertebrate haemoglobin
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout
Nature 581, 388-389 (2020)
doi: https://doi.org/10.1038/d41586-020-01287-8
References
Berenbrink, M., Koldkjær, P., Kepp, O. & Cossins, A. R. Science 307, 1752–1757 (2005).
Natarajan, C. et al. Science 354, 336–339 (2016).
Perutz, M. F. Mol. Biol. Evol. 1, 1–28 (1983).
Pillai, A. S. et al. Nature 581, 480–485 (2020).
Antonini, E. & Brunori, M. Hemoglobin and Myoglobin in Their Reactions with Ligands 21 (North-Holland, 1971).
Benner, S. A. in Ancestral Sequence Reconstruction (ed. Liberles, D. A.) Ch. 1, 3–19 (Oxford Univ. Press, 2007).
Krogh, A. & Leitch, I. J. Physiol. (Lond.) 52, 288–300 (1919).
Storz, J. F. Hemoglobin: Insights into Protein Structure, Function, and Evolution (Oxford Univ. Press, 2019).
Imai, K. Allosteric Effects in Haemoglobin (Cambridge Univ. Press, 1982).
Schachat, S. R. et al. Proc. R. Soc. B 285, 20172631 (2018).
Rund, D. & Rachmilewitz, E. N. Engl. J. Med. 353, 1135–1146 (2005).
Mirceta, S. et al. Science 340, 1234192 (2013).
Zuckerkandl, E. in Ancestral Sequence Reconstruction (ed. Liberles, D. A.) x–xi (Oxford Univ. Press, 2007).

 Read the paper: Origin of complexity in haemoglobin evolution
Read the paper: Origin of complexity in haemoglobin evolution
 A ratchet for protein complexity
A ratchet for protein complexity
 Network evolution hinges on history
Network evolution hinges on history