- NEWS AND VIEWS
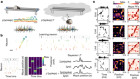
Neuroimaging results altered by varying analysis pipelines
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout
Nature 582, 36-37 (2020)
doi: https://doi.org/10.1038/d41586-020-01282-z
References
Carp, J. NeuroImage 63, 289–300 (2012).
Botvinik-Nezer, R. et al. Nature 582, 84–88 (2020).
Carp, J. Front. Neurosci. 6, 149 (2012).
Simmons, J. P., Nelson, L. D. & Simonsohn, U. Psychol. Sci. 22, 1359–1366 (2011).
Strother, S. C. et al. NeuroImage 15, 747–771 (2002).
Churchill, N. W. et al. Hum. Brain Mapp. 33, 609–627 (2012).
Woo, C. W., Chang, L. J., Lindquist, M. A. & Wager, T. D. Nature Neurosci. 20, 365 (2017).

 Read the paper: Variability in the analysis of a single neuroimaging dataset by many teams
Read the paper: Variability in the analysis of a single neuroimaging dataset by many teams
 Experimental mismatch in neural circuits
Experimental mismatch in neural circuits
 Occasional errors can benefit coordination
Occasional errors can benefit coordination