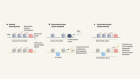
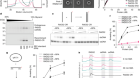
- NEWS AND VIEWS
Limitless translation limits translation
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout
Nature 553, 289-290 (2018)
doi: https://doi.org/10.1038/d41586-017-08785-2
References
Yordanova, M. M. et al. Nature 553, 356–360 (2018).
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. S. & Weissman, J. S. Science 324, 218–223 (2009).
Van Damme, P., Gawron, D., Van Criekinge, W. & Menschaert, G. Mol. Cell. Proteomics 13, 1245–1261 (2014).
Dunn, J. G., Foo, C. K., Belletier, N. G., Gavis, E. R. & Weissman, J. S. eLife 2, e01179 (2013).
Smith, L. M., Kelleher, N. L. & The Consortium for Top Down Proteomics Nature Methods 10, 186–187 (2013).
Law, G. L., Raney, A., Heusner, C. & Morris, D. R. J. Biol. Chem. 276, 38036–38043 (2001).
Doronina, V. A. et al. Mol. Cell. Biol. 28, 4227–4239 (2008).
Arribere, J. A. et al. Nature 534, 719–723 (2016).
MacDonald, C. T., Gibbs, J. H. & Pipkin, A. C. Biopolymers 6, 1–25 (1968).
Ferrin, M. A. & Subramaniam, A. R. eLife 6, e23629 (2017).
Wang, C., Han, B., Zhou, R. & Zhuang, X. Cell 165, 990–1001 (2016).

 Read the paper: AMD1 mRNA employs ribosome stalling as a mechanism for molecular memory formation
Read the paper: AMD1 mRNA employs ribosome stalling as a mechanism for molecular memory formation
 Unconventional translation in cancer
Unconventional translation in cancer
 Entry signals control development
Entry signals control development