- NEWS AND VIEWS
How chromosomes unite
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout
Nature 551, 568-569 (2017)
doi: https://doi.org/10.1038/d41586-017-07439-7
References
Samwer, M. et al. Cell 170, 956–972 (2017).
Hatch, E. M., Fischer, A. H., Deerinck, T. J. & Hetzer, M. W. Cell 154, 47–60 (2013).
Zhang, C.-Z. et al. Nature 522, 179–184 (2015).
Gekara, N. O. J. Cell Biol. 216, 2999–3001 (2017).
Margalit, A., Segura-Totten, M., Gruenbaum, Y. & Wilson, K. L. Proc. Natl Acad. Sci. USA 102, 3290–3295 (2005).
Gorjánácz, M. et al. EMBO J. 26, 132–143 (2007).
Lee, M. S. & Craigie, R. Proc. Natl Acad. Sci. USA 95, 1528–1533 (1998).
Umland, T. C., Wei, S.-Q., Craigie, R. & Davies, D. R. Biochemistry 39, 9130–9138 (2000).
Shibata, Y., Voeltz, G. K. & Rapoport, T. A. Cell 126, 435–439 (2006).
Segura-Totten, M., Kowalski, A. K., Craigie, R. & Wilson, K. L. J. Cell Biol. 158, 475–485 (2002).
Bradley, C. M., Ronning, D. R., Ghirlando, R., Craigie, R. & Dyda, F. Nature Struct. Mol. Biol. 12, 935–936 (2005).
Zheng, R., Ghirlando, R., Lee, M. S., Mizuuchi, K., Krause, M. & Craigie, R. Proc. Natl Acad. Sci. USA 97, 8997–9002 (2000).
Montes de Oca, R., Lee, K. K. & Wilson, K. L. J. Biol. Chem. 280, 42252–42262 (2005).
Haraguchi, T. et al. J. Cell Sci. 121, 2540–2554 (2008).
Wong, X., Luperchio, T. R. & Reddy, K. L. Curr. Opin. Cell Biol. 28, 105–120 (2014).
Kim, S. H. et al. J. Pharmacol. Exp. Ther. 352, 175–184 (2015).
Gorjánácz, M. Nucleus 5, 47–55 (2014).
Jamin, A. & Wiebe, M. S. Curr. Opin. Cell Biol. 34, 61–68 (2015).

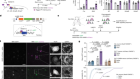
 Organelle formation from scratch
Organelle formation from scratch
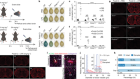
 A sticky problem for chromosomes
A sticky problem for chromosomes